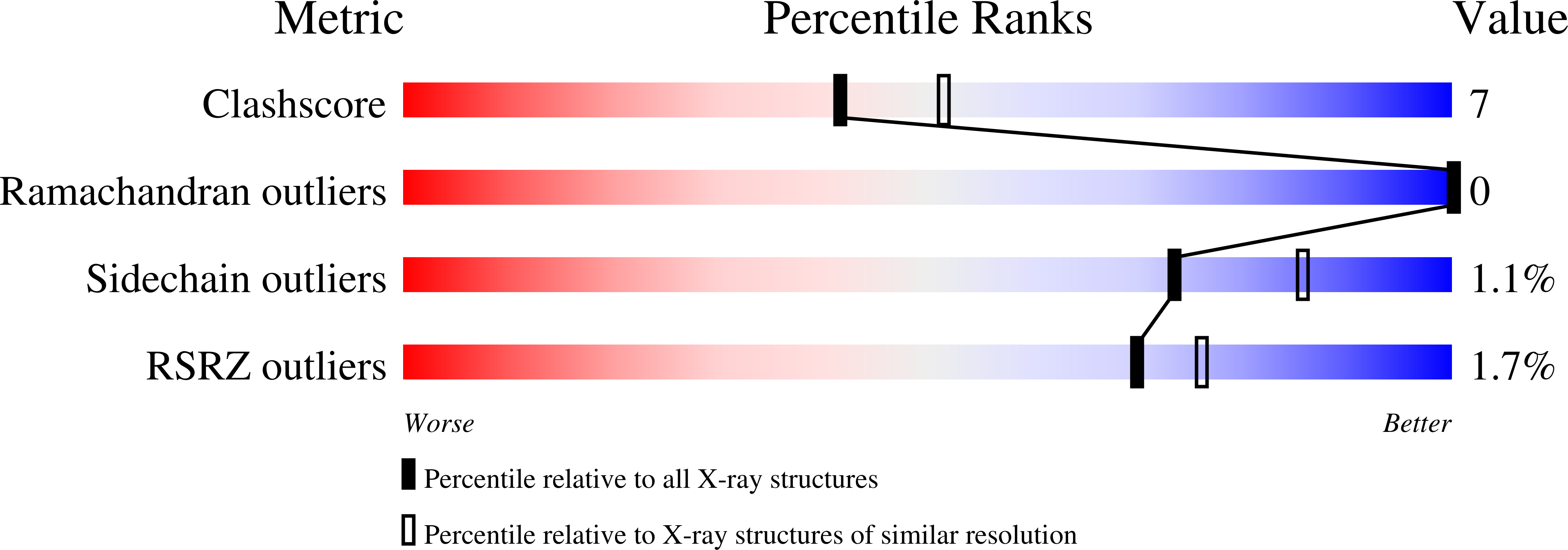
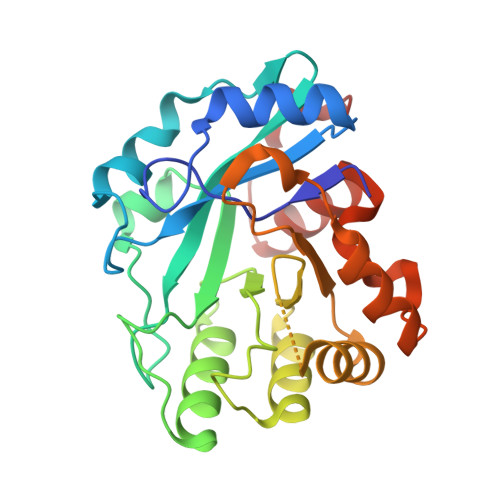
Crystal structure of the Escherichia coli YcdX protein reveals a trinuclear zinc active site
Teplyakov, A., Obmolova, G., Khil, P.P., Howard, A.J., Camerini-Otero, R.D., Gilliland, G.L.(2003) Proteins 51: 315-318
- PubMed: 12661000
- DOI: https://doi.org/10.1002/prot.10352
- Primary Citation of Related Structures:
1M65, 1M68
Organizational Affiliation:
Center for Advanced Research in Biotechnology, University of Maryland Biotechnology Institute and the National Institute of Standards and Technology, Rockville, Maryland 20850, USA.