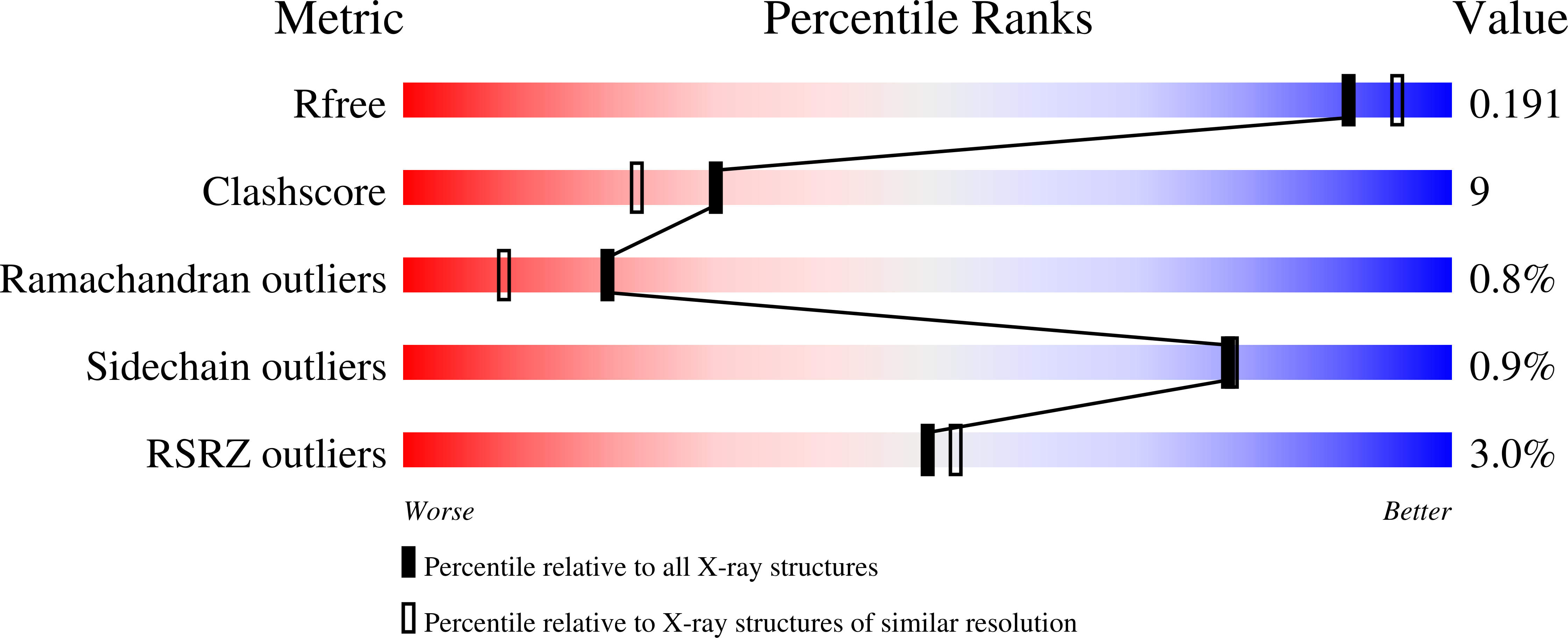
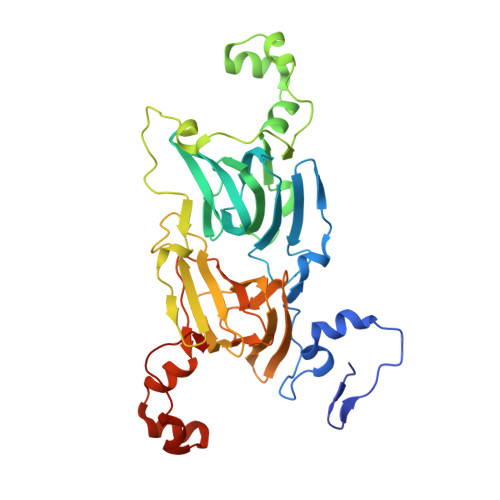
Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 A resolution.
Anand, R., Dorrestein, P.C., Kinsland, C., Begley, T.P., Ealick, S.E.(2002) Biochemistry 41: 7659-7669
- PubMed: 12056897
- DOI: https://doi.org/10.1021/bi0200965
- Primary Citation of Related Structures:
1J58, 1L3J - PubMed Abstract:
Oxalate decarboxylase is a manganese-dependent enzyme that catalyzes the conversion of oxalate to formate and carbon dioxide. We have determined the structure of oxalate decarboxylase from Bacillus subtilis at 1.75 A resolution in the presence of formate. The structure reveals a hexamer with 32-point symmetry in which each monomer belongs to the cupin family of proteins. Oxalate decarboxylase is further classified as a bicupin because it contains two cupin folds, possibly resulting from gene duplication. Each oxalate decarboxylase cupin domain contains one manganese binding site. Each of the oxalate decarboxylase domains is structurally similar to oxalate oxidase, which catalyzes the manganese-dependent oxidative decarboxylation of oxalate to carbon dioxide and hydrogen peroxide. Amino acid side chains in the two metal binding sites of oxalate decarboxylase and the metal binding site of oxalate oxidase are very similar. Four manganese binding residues (three histidines and one glutamate) are conserved as well as a number of hydrophobic residues. The most notable difference is the presence of Glu333 in the metal binding site of the second cupin domain of oxalate decarboxylase. We postulate that this domain is responsible for the decarboxylase activity and that Glu333 serves as a proton donor in the production of formate. Mutation of Glu333 to alanine reduces the catalytic activity by a factor of 25. The function of the other domain in oxalate decarboxylase is not yet known.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Cornell University, Ithaca, New York 14853, USA.