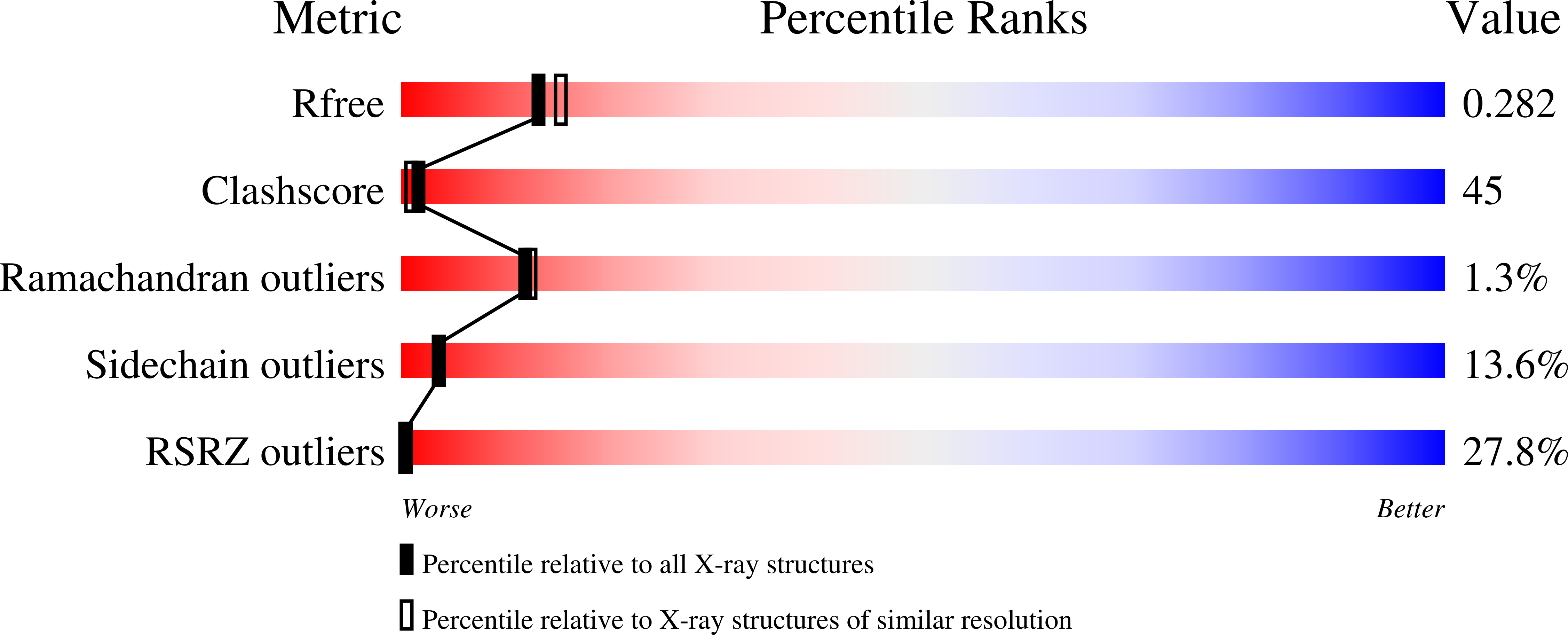
Thermolysin in the absence of substrate has an open conformation.
Hausrath, A.C., Matthews, B.W.(2002) Acta Crystallogr D Biol Crystallogr 58: 1002-1007
- PubMed: 12037302
- DOI: https://doi.org/10.1107/s090744490200584x
- Primary Citation of Related Structures:
1L3F - PubMed Abstract:
The bacterial neutral proteases have been proposed to undergo hinge-bending during their catalytic cycle. However, in thermolysin, the prototypical member of the family, no significant conformational change has been observed. The structure of thermolysin has now been determined in a new crystal form that for the first time shows the enzyme in the absence of a ligand bound in the active site. This is shown to be an 'open' form of the enzyme. The relative orientation of the two domains that define the active-site cleft differ by a 5 degrees rotation relative to their positions in the previously studied ligand-bound 'closed' form. Based on structural comparisons, kinetic studies on mutants and molecular-dynamics simulations, Gly78 and Gly135-Gly136 have previously been suggested as two possible hinge regions. Comparison of the 'open' and 'closed' structures suggests that neither of the proposed hinge regions completely accounts for the observed displacement. The concerted movement of a group of side chains suggested to be associated with the hinge-bending motion is, however, confirmed.
Organizational Affiliation:
Institute of Molecular Biology, Howard Hughes Medical Institute and Department of Physics, 1229 University of Oregon, Eugene, OR 97403-1229, USA.