Solution structure of a luteoviral P1-P2 frameshifting mRNA pseudoknot
Nixon, P.L., Rangan, A., Kim, Y.-G., Rich, A., Hoffman, D.W., Hennig, M., Giedroc, D.P.(2002) J Mol Biol 322: 621-633
- PubMed: 12225754
- DOI: https://doi.org/10.1016/s0022-2836(02)00779-9
- Primary Citation of Related Structures:
1KPY, 1KPZ - PubMed Abstract:
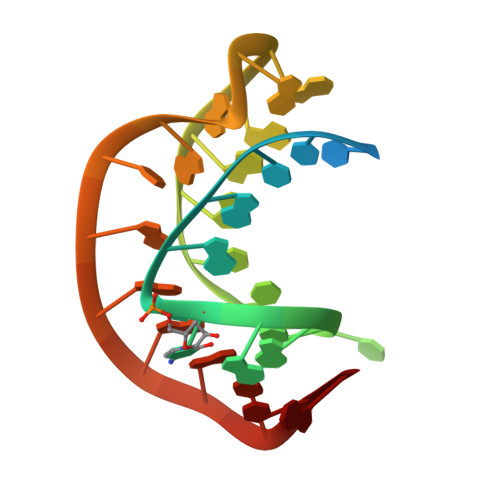
A hairpin-type messenger RNA pseudoknot from pea enation mosaic virus RNA1 (PEMV-1) regulates the efficiency of programmed -1 ribosomal frameshifting. The solution structure and 15N relaxation rates reveal that the PEMV-1 pseudoknot is a compact-folded structure composed almost entirely of RNA triple helix. A three nucleotide reverse turn in loop 1 positions a protonated cytidine, C(10), in the correct orientation to form an A((n-1)).C(+).G-C(n) major groove base quadruple, like that found in the beet western yellows virus pseudoknot and the hepatitis delta virus ribozyme, despite distinct structural contexts. A novel loop 2-loop 1 A.U Hoogsteen base-pair stacks on the C(10)(+).G(28) base-pair of the A(12).C(10)(+).G(28)-C(13) quadruple and forms a wedge between the pseudoknot stems stabilizing a bent and over-rotated global conformation. Substitution of key nucleotides that stabilize the unique conformation of the PEMV-1 pseudoknot greatly reduces ribosomal frameshifting efficacy.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Center for Advanced Biomolecular Research, 2128 TAMU, Texas A&M University, 77843-2128, College Station, TX, USA.