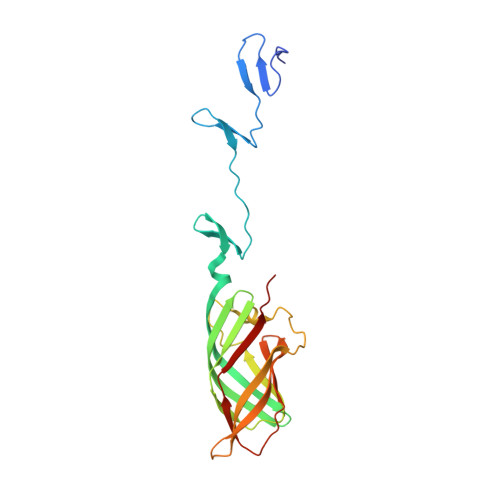
Crystal structure of reovirus attachment protein sigma1 reveals evolutionary relationship to adenovirus fiber.
Chappell, J.D., Prota, A.E., Dermody, T.S., Stehle, T.(2002) EMBO J 21: 1-11
- PubMed: 11782420
- DOI: https://doi.org/10.1093/emboj/21.1.1
- Primary Citation of Related Structures:
1KKE - PubMed Abstract:
Reovirus attaches to cellular receptors with the sigma1 protein, a fiber-like molecule protruding from the 12 vertices of the icosahedral virion. The crystal structure of a receptor-binding fragment of sigma1 reveals an elongated trimer with two domains: a compact head with a new beta-barrel fold and a fibrous tail containing a triple beta-spiral. Numerous structural and functional similarities between reovirus sigma1 and the adenovirus fiber suggest an evolutionary link in the receptor-binding strategies of these two viruses. A prominent loop in the sigma1 head contains a cluster of residues that are conserved among reovirus serotypes and are likely to form a binding site for junction adhesion molecule, an integral tight junction protein that serves as a reovirus receptor. The fibrous tail is mainly responsible for sigma1 trimer formation, and it contains a highly flexible region that allows for significant movement between the base of the tail and the head. The architecture of the trimer interface and the observed flexibility indicate that sigma1 is a metastable structure poised to undergo conformational changes upon viral attachment and cell entry.
Organizational Affiliation:
Department of Pediatrics, Vanderbilt University School of Medicine, Nashville, TN 37232, USA.