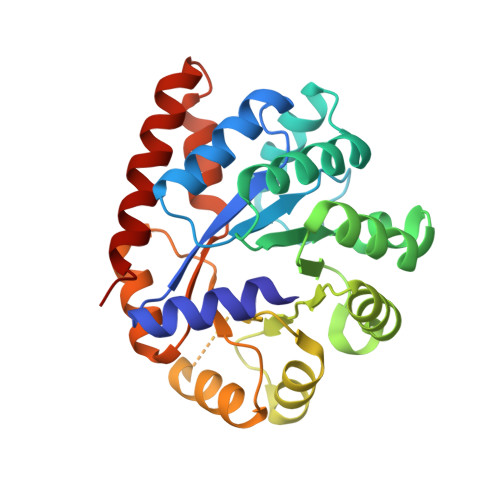
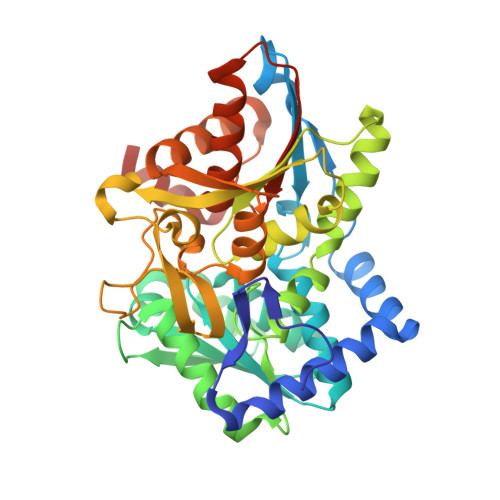
Crystal structure of the beta Ser178--> Pro mutant of tryptophan synthase. A "knock-out" allosteric enzyme.
Weyand, M., Schlichting, I., Herde, P., Marabotti, A., Mozzarelli, A.(2002) J Biol Chem 277: 10653-10660
- PubMed: 11756454
- DOI: https://doi.org/10.1074/jbc.M111031200
- Primary Citation of Related Structures:
1K7X, 1K8Y, 1K8Z - PubMed Abstract:
The catalytic activity of the pyridoxal 5'-phosphate-dependent tryptophan synthase alpha(2)beta(2) complex is allosterically regulated. The hydrogen bond between the helix betaH6 residue betaSer(178) and the loop alphaL6 residue Gly(181) was shown to be critical in ligand-induced intersubunit signaling, with the alpha-beta communication being completely lost in the mutant betaSer(178) --> Pro (Marabotti, A., De Biase, D., Tramonti, A., Bettati, S., and Mozzarelli, A. (2001) J. Biol. Chem. 276, 17747-17753). The structural basis of the impaired allosteric regulation was investigated by determining the crystal structures of the mutant betaSer(178) --> Pro in the absence and presence of the alpha-subunit ligands indole-3-acetylglycine and glycerol 3-phosphate. The mutation causes local and distant conformational changes especially in the beta-subunit. The ligand-free structure exhibits larger differences at the N-terminal part of helix betaH6, whereas the enzyme ligand complexes show differences at the C-terminal side. In contrast to the wild-type enzyme loop alphaL6 remains in an open conformation even in the presence of alpha-ligands. This effects the equilibrium between active and inactive conformations of the alpha-active site, altering k(cat) and K(m), and forms the structural basis for the missing allosteric communication between the alpha- and beta-subunits.
Organizational Affiliation:
Max-Planck-Institut für Molekulare Physiologie, Abteilung für Physikalische Biochemie, D-44227 Dortmund, Germany.