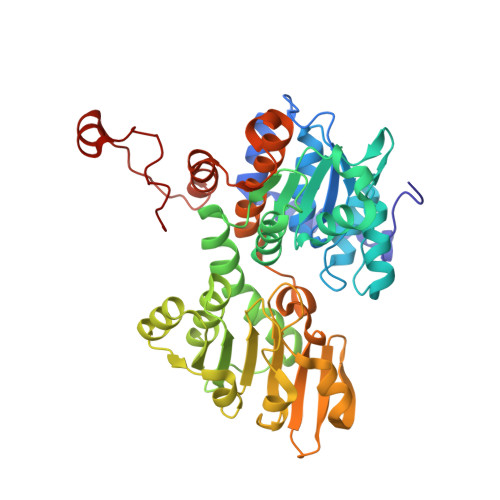
Inhibition of S-adenosylhomocysteine hydrolase by acyclic sugar adenosine analogue D-eritadenine. Crystal structure of S-adenosylhomocysteine hydrolase complexed with D-eritadenine.
Huang, Y., Komoto, J., Takata, Y., Powell, D.R., Gomi, T., Ogawa, H., Fujioka, M., Takusagawa, F.(2002) J Biol Chem 277: 7477-7482
- PubMed: 11741948
- DOI: https://doi.org/10.1074/jbc.M109187200
- Primary Citation of Related Structures:
1K0U - PubMed Abstract:
D-eritadenine (DEA) is a potent inhibitor (IC(50) = 7 nm) of S-adenosyl-l-homocysteine hydrolase (AdoHcyase). Unlike cyclic sugar Ado analogue inhibitors, including mechanism-based inhibitors, DEA is an acyclic sugar Ado analogue, and the C2' and C3' have opposite chirality to those of the cyclic sugar Ado inhibitors. Crystal structures of DEA alone and in complex with AdoHcyase have been determined to elucidate the DEA binding scheme to AdoHcyase. The DEA-complexed structure has been analyzed by comparing it with two structures of AdoHcyase complexed with cyclic sugar Ado analogues. The DEA-complexed structure has a closed conformation, and the DEA is located near the bound NAD(+). However, a UV absorption measurement shows that DEA is not oxidized by the bound NAD(+), indicating that the open-closed conformational change of AdoHcyase is due to the substrate/inhibitor binding, not the oxidation state of the bound NAD. The adenine ring of DEA is recognized by four essential hydrogen bonds as observed in the cyclic sugar Ado complexes. The hydrogen bond network around the acyclic sugar moiety indicates that DEA is more tightly connected to the protein than the cyclic sugar Ado analogues. The C3'-H of DEA is pointed toward C4 of the bound NAD(+) (C3'...C4 = 3.7 A), suggesting some interaction between DEA and NAD(+). By placing DEA into the active site of the open structure, the major forces to stabilize the closed conformation of AdoHcyase are identified as the hydrogen bonds between the backbone of His-352 and the adenine ring, and the C3'-H...C4 interaction. DEA has been believed to be an inactivator of AdoHcyase, but this study indicates that DEA is a reversible inhibitor. On the basis of the complexed structure, selective inhibitors of AdoHcyase have been designed.
Organizational Affiliation:
Department of Molecular Biosciences, University of Kansas, 1200 Sunnyside Avenue, Lawrence, KS 66045-7534, USA.