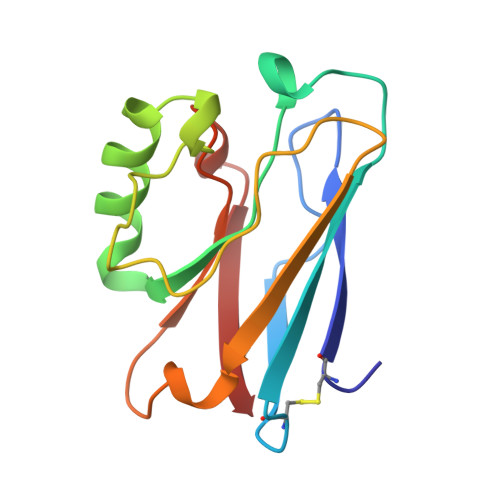
Crystallization and preliminary crystallographic studies of the crystals of the azurin Pseudomonas fluorescens.
Zhu, D.W., Dahms, T., Willis, K., Szabo, A.G., Lee, X.(1994) Arch Biochem Biophys 308: 469-470
- PubMed: 8109977
- DOI: https://doi.org/10.1006/abbi.1994.1066
- Primary Citation of Related Structures:
1JOI - PubMed Abstract:
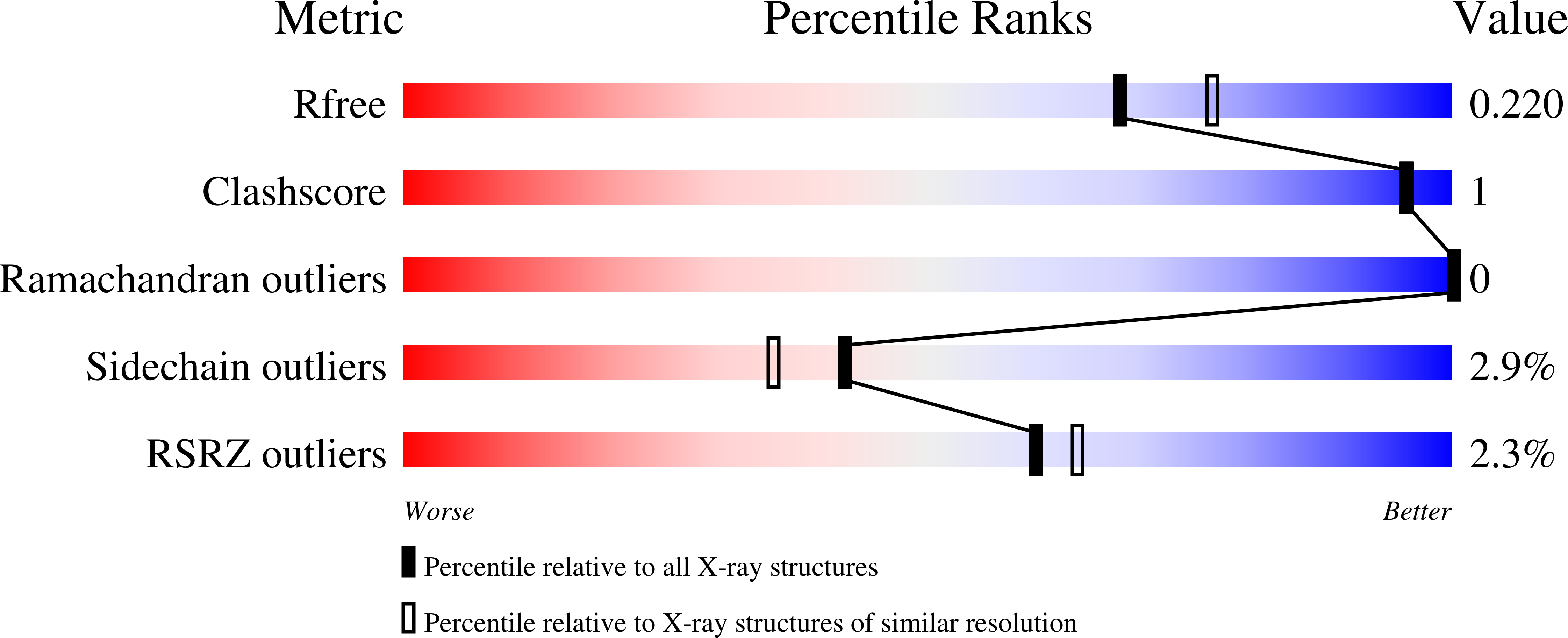
The Azurin Pseudomones fluorescens has been crystallized in the presence of ammonium sulfate and Tris buffer at pH 7.5. The crystals diffract to 2.05 A using a FAST system. The space group is P2(1)2(1)2(1) with a = 31.95, b = 43.78, and c = 78.81 A.
Organizational Affiliation:
MRC Group in Molecular Endocrinology, CHUL Research Center, Ste-Foy, Québec, Canada.