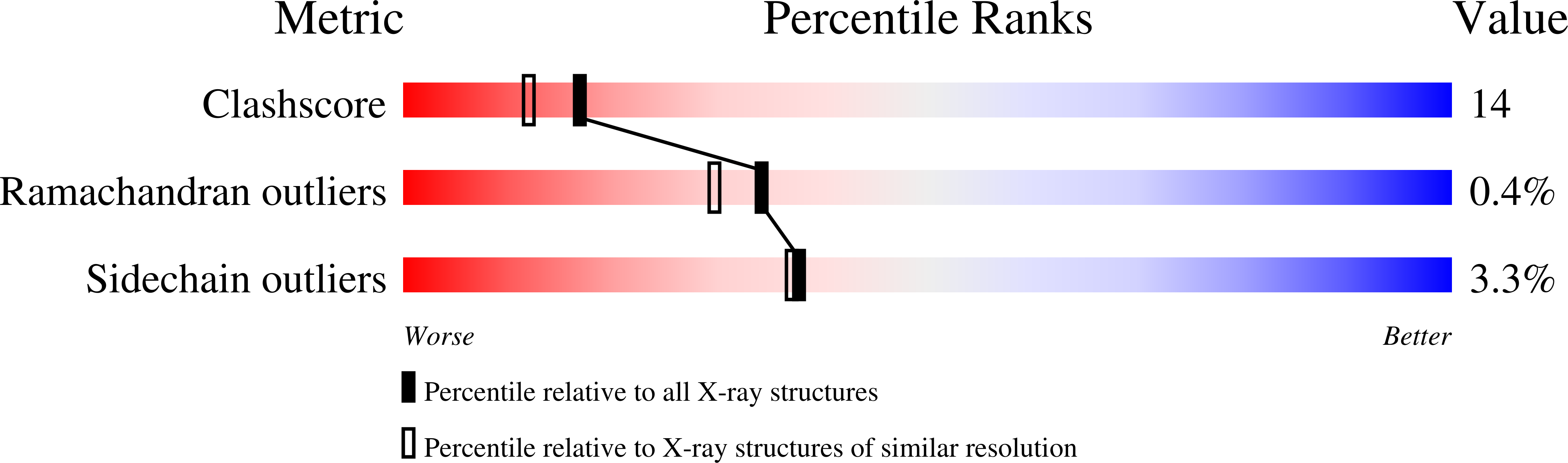Chicken avidin exhibits pseudo-catalytic properties. Biochemical, structural, and electrostatic consequences.
Huberman, T., Eisenberg-Domovich, Y., Gitlin, G., Kulik, T., Bayer, E.A., Wilchek, M., Livnah, O.(2001) J Biol Chem 276: 32031-32039
- PubMed: 11395489
- DOI: https://doi.org/10.1074/jbc.M102018200
- Primary Citation of Related Structures:
1I9H, 1IJ8 - PubMed Abstract:
Avidin and its bacterial analogue streptavidin exhibit similarly high affinities toward the vitamin biotin. The extremely high affinity of these two proteins has been utilized as a powerful tool in many biotechnological applications. Although avidin and streptavidin have similar tertiary and quaternary structures, they differ in many of their properties. Here we show that avidin enhances the alkaline hydrolysis of biotinyl p-nitrophenyl ester, whereas streptavidin protects this reaction even under extreme alkaline conditions (pH > 12). Unlike normal enzymatic catalysis, the hydrolysis reaction proceeds as a single cycle with no turnover because of the extremely high affinity of the protein for one of the reaction products (i.e. free biotin). The three-dimensional crystal structures of avidin (2 A) and streptavidin (2.4 A) complexed with the amide analogue, biotinyl p-nitroanilide, as a model for the p-nitrophenyl ester, revealed structural insights into the factors that enhance or protect the hydrolysis reaction. The data demonstrate that several molecular features of avidin are responsible for the enhanced hydrolysis of biotinyl p-nitrophenyl ester. These include the nature of a decisive flexible loop, the presence of an obtrusive arginine 114, and a newly formed critical interaction between lysine 111 and the nitro group of the substrate. The open conformation of the loop serves to expose the substrate to the solvent, and the arginine shifts the p-nitroanilide moiety toward the interacting lysine, which increases the electron withdrawing characteristics and consequent electrophilicity of the carbonyl group of the substrate. Streptavidin lacked such molecular properties, and analogous interactions with the substrate were consequently absent. The information derived from these structures may provide insight into the action of artificial protein catalysts and the evolution of catalytic sites in general.
Organizational Affiliation:
Department of Biological Chemistry, The Institute of Life Sciences, Wolfson Centre for Applied Structural Biology, Hebrew University of Jerusalem, Givat Ram, Jerusalem 91904, Israel.