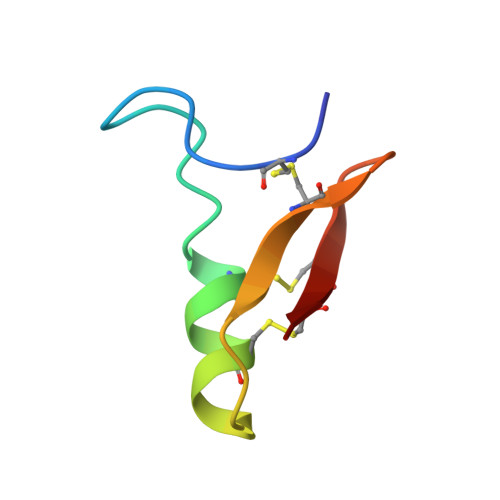
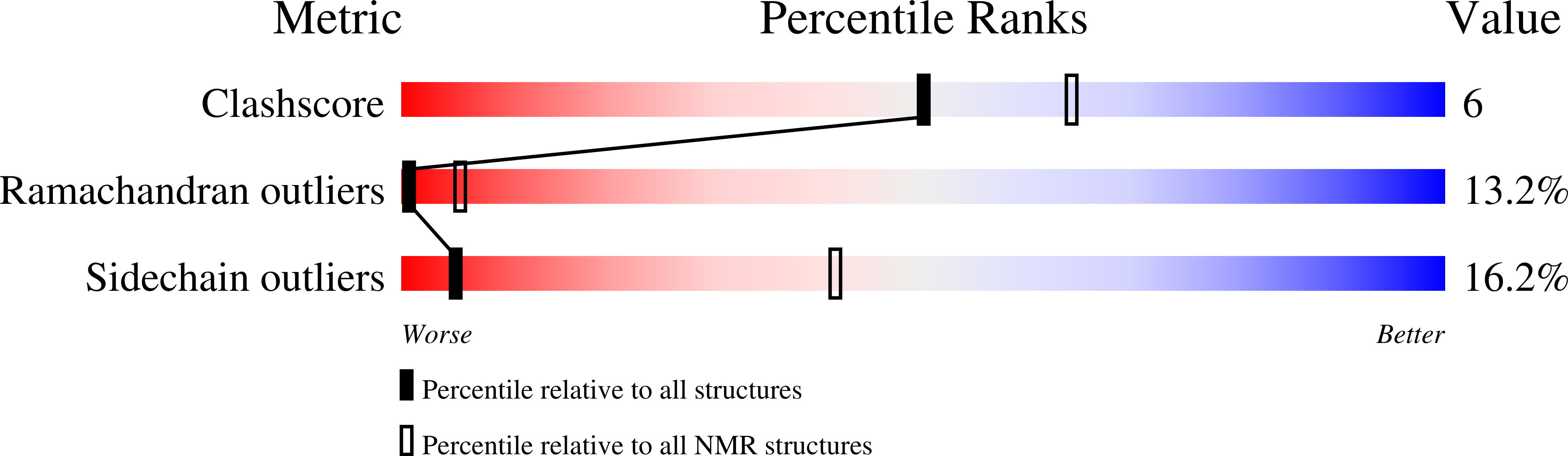Refined three-dimensional solution structure of insect defensin A.
Cornet, B., Bonmatin, J.M., Hetru, C., Hoffmann, J.A., Ptak, M., Vovelle, F.(1995) Structure 3: 435-448
- PubMed: 7663941
- DOI: https://doi.org/10.1016/s0969-2126(01)00177-0
- Primary Citation of Related Structures:
1ICA - PubMed Abstract:
Insect defensin A is a basic 4 kDa protein secreted by Phormia terranovae larvae in response to bacterial challenges or injuries. Previous biological tests suggest that the bacterial cytoplasmic membrane is the target of defensin A. The structural study of this protein is the first step towards establishing a structure-activity relationship and forms the basis for understanding its antibiotic activity at the molecular level. We describe a refined model of the three-dimensional structure of defensin A derived from an extensive analysis of 786 inter-proton nuclear Overhauser effects. The backbone fold involves an N-terminal loop and an alpha-helical fragment followed by an antiparallel beta-structure. The helix and the beta-structure are connected by two of the three disulphide bridges present in defensin A, forming a so-called 'cysteine-stabilized alpha beta' (CS alpha beta) motif. The N-terminal loop, which is locally well defined, can occupy different positions with respect to the other moieties of the molecule. The CS alpha beta motif, which forms the core of the defensin A structure, appears to be a common organization for several families of small proteins with toxic properties. The distribution of amino acid side chains in the protein structure creates several hydrophobic or hydrophilic patches. This leads us to propose that the initial step in the action of positively charged defensin A molecules with cytoplasmic membranes may involve interactions with acidic phospholipids.
Organizational Affiliation:
Centre de Biophysique Moléculaire (CNRS), Orléans, France.