Corepressor-induced organization and assembly of the biotin repressor: a model for allosteric activation of a transcriptional regulator.
Weaver, L.H., Kwon, K., Beckett, D., Matthews, B.W.(2001) Proc Natl Acad Sci U S A 98: 6045-6050
- PubMed: 11353844
- DOI: https://doi.org/10.1073/pnas.111128198
- Primary Citation of Related Structures:
1HXD - PubMed Abstract:
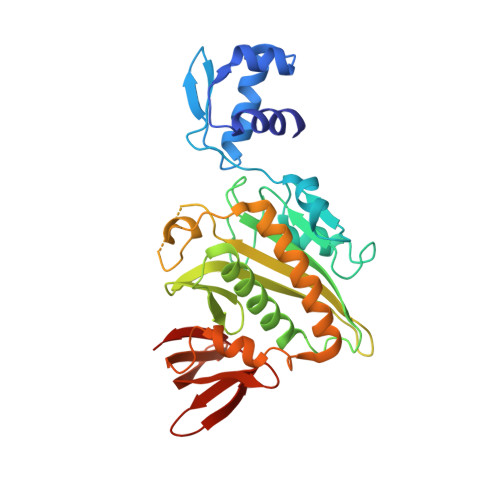
The Escherichia coli biotin repressor binds to the biotin operator to repress transcription of the biotin biosynthetic operon. In this work, a structure determined by x-ray crystallography of a complex of the repressor bound to biotin, which also functions as an activator of DNA binding by the biotin repressor (BirA), is described. In contrast to the monomeric aporepressor, the complex is dimeric with an interface composed in part of an extended beta-sheet. Model building, coupled with biochemical data, suggests that this is the dimeric form of BirA that binds DNA. Segments of three surface loops that are disordered in the aporepressor structure are located in the interface region of the dimer and exhibit greater order than was observed in the aporepressor structure. The results suggest that the corepressor of BirA causes a disorder-to-order transition that is a prerequisite to repressor dimerization and DNA binding.
Organizational Affiliation:
Institute for Molecular Biology, Howard Hughes Medical Institute and Department of Physics, 1229 University of Oregon, Eugene, OR 97403-1229, USA. brian@uoxray.uoregon.edu