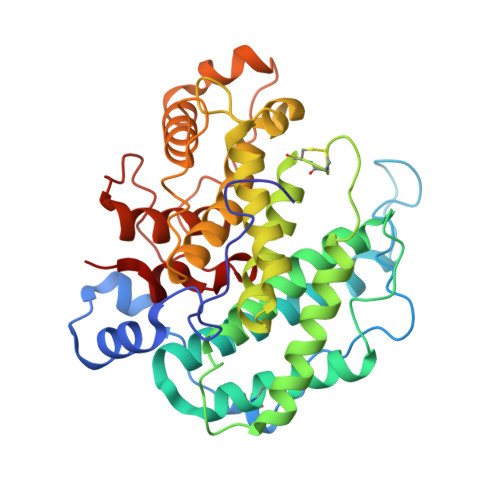
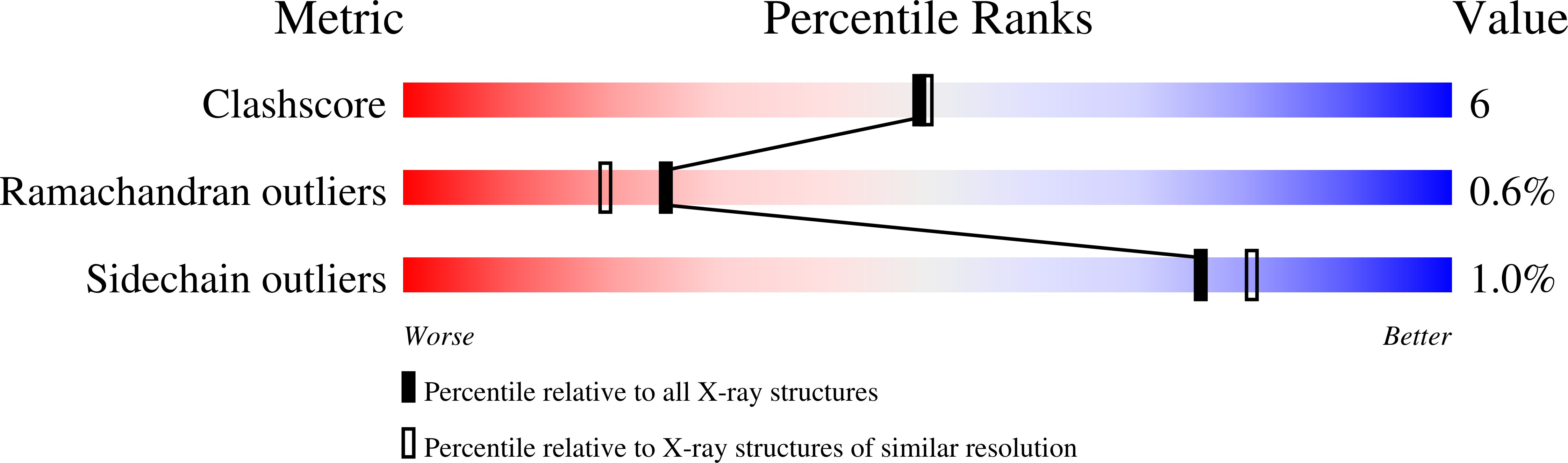Crystal structure of alginate lyase A1-III complexed with trisaccharide product at 2.0 A resolution.
Yoon, H.J., Hashimoto, W., Miyake, O., Murata, K., Mikami, B.(2001) J Mol Biol 307: 9-16
- PubMed: 11243798
- DOI: https://doi.org/10.1006/jmbi.2000.4509
- Primary Citation of Related Structures:
1HV6 - PubMed Abstract:
The structure of A1-III from a Sphingomonas species A1 complexed with a trisaccharide product (4-deoxy-l-erythro-hex-4-enepyranosyluronate-mannuronate-mannuronic acid) was determined by X-ray crystallography at 2.0 A with an R-factor of 0.16. The final model of the complex form comprising 351 amino acid residues, 245 water molecules, one sulfate ion and one trisaccharide product exhibited a C(alpha) r.m.s.d. value of 0.154 A with the reported apo form of the enzyme. The trisaccharide was bound in the active cleft at subsites -3 approximately -1 from the non-reducing end by forming several hydrogen bonds and van der Waals interactions with protein atoms. The catalytic residue was estimated to be Tyr246, which existed between subsites -1 and +1 based on a mannuronic acid model oriented at subsite +1.
Organizational Affiliation:
Research Institute for Food Science, Kyoto University, Uji Kyoto 611-0011, Japan.