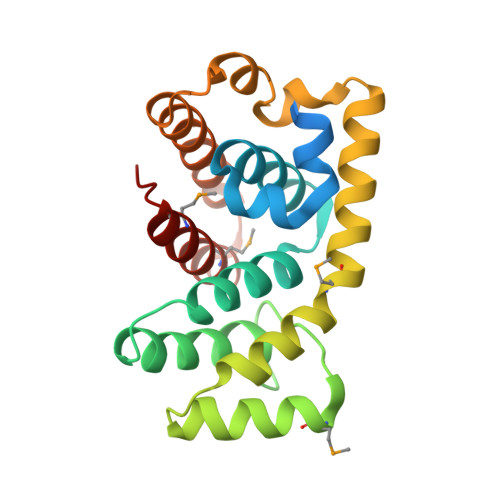
Structure of the RGS-like domain from PDZ-RhoGEF: linking heterotrimeric g protein-coupled signaling to Rho GTPases.
Longenecker, K.L., Lewis, M.E., Chikumi, H., Gutkind, J.S., Derewenda, Z.S.(2001) Structure 9: 559-569
- PubMed: 11470431
- DOI: https://doi.org/10.1016/s0969-2126(01)00620-7
- Primary Citation of Related Structures:
1HTJ - PubMed Abstract:
The multidomain PDZ-RhoGEF is one of many known guanine nucleotide exchange factors that upregulate Rho GTPases. PDZ-RhoGEF and related family members play a critical role in a molecular signaling pathway from heterotrimeric G protein-coupled receptors to Rho proteins. A approximately 200 residue RGS-like (RGSL) domain in PDZ-RhoGEF and its homologs is responsible for the direct association with Galpha12/13 proteins. To better understand structure-function relationships, we initiated crystallographic studies of the RGSL domain from human PDZ-RhoGEF.
Organizational Affiliation:
Department of Molecular Physiology and Biological Physics, University of Virginia, P.O. Box 800736, Charlottesville, VA 22908, USA.