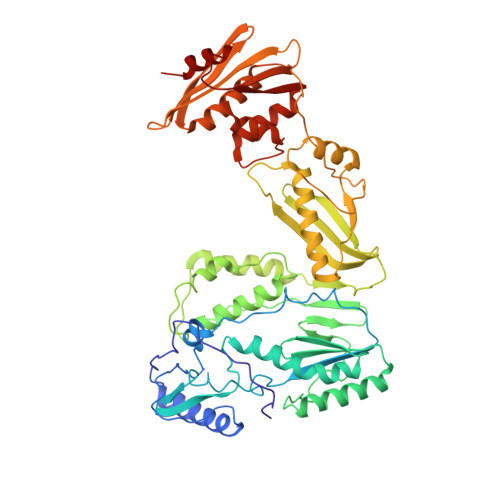
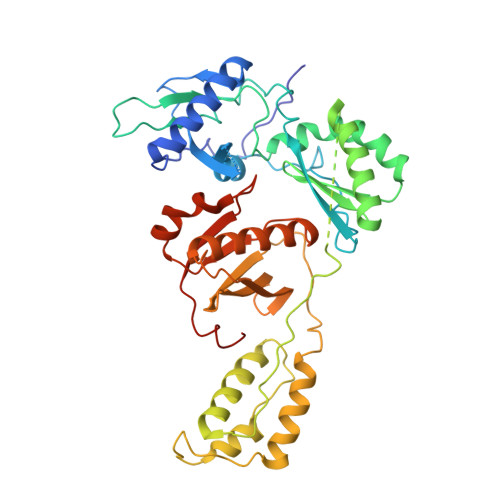
The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1.
Rodgers, D.W., Gamblin, S.J., Harris, B.A., Ray, S., Culp, J.S., Hellmig, B., Woolf, D.J., Debouck, C., Harrison, S.C.(1995) Proc Natl Acad Sci U S A 92: 1222-1226
- PubMed: 7532306
- DOI: https://doi.org/10.1073/pnas.92.4.1222
- Primary Citation of Related Structures:
1HMV - PubMed Abstract:
The crystal structure of the reverse transcriptase (RT) from the type 1 human immunodeficiency virus has been determined at 3.2-A resolution. Comparison with complexes between RT and the polymerase inhibitor Nevirapine [Kohlstaedt, L.A., Wang, J., Friedman, J.M., Rice, P.A. & Steitz, T.A. (1992) Science 256, 1783-1790] and between RT and an oligonucleotide [Jacobo-Molina, A., Ding, J., Nanni, R., Clark, A. D., Lu, X., Tantillo, C., Williams, R. L., Kamer, G., Ferris, A. L., Clark, P., Hizi, A., Hughes, S. H. & Arnold, E. (1993) Proc. Natl. Acad. Sci. USA 90, 6320-6324] reveals changes associated with ligand binding. The enzyme is a heterodimer (p66/p51), with domains labeled "fingers," "thumb," "palm," and "connection" in both subunits, and a ribonuclease H domain in the larger subunit only. The most striking difference between RT and both complex structures is the change in orientation of the p66 thumb (approximately 33 degrees rotation). Smaller shifts relative to the core of the molecule were also found in other domains, including the p66 fingers and palm, which contain the polymerase active site. Within the polymerase catalytic region itself, there are no rearrangements between RT and the RT/DNA complex. In RT/Nevirapine, the drug binds in the p66 palm near the polymerase active site, a region that is well-packed hydrophobic core in the unliganded enzyme. Room for the drug is provided by movement of a small beta-sheet within the palm domain of the Nevirapine complex. The rearrangement within the palm and thumb, as well as domain shifts relative to the enzyme core, may prevent correct placement of the oligonucleotide substrate when the drug is bound.
Organizational Affiliation:
Department of Molecular and Cellular Biology, Harvard University, Cambridge, MA 02138.