Crystal Structure of the Bacterial Cell-Division Inhibitor Minc
Cordell, S.C., Anderson, R.E., Lowe, J.(2001) EMBO J 20: 2454
- PubMed: 11350934
- DOI: https://doi.org/10.1093/emboj/20.10.2454
- Primary Citation of Related Structures:
1HF2 - PubMed Abstract:
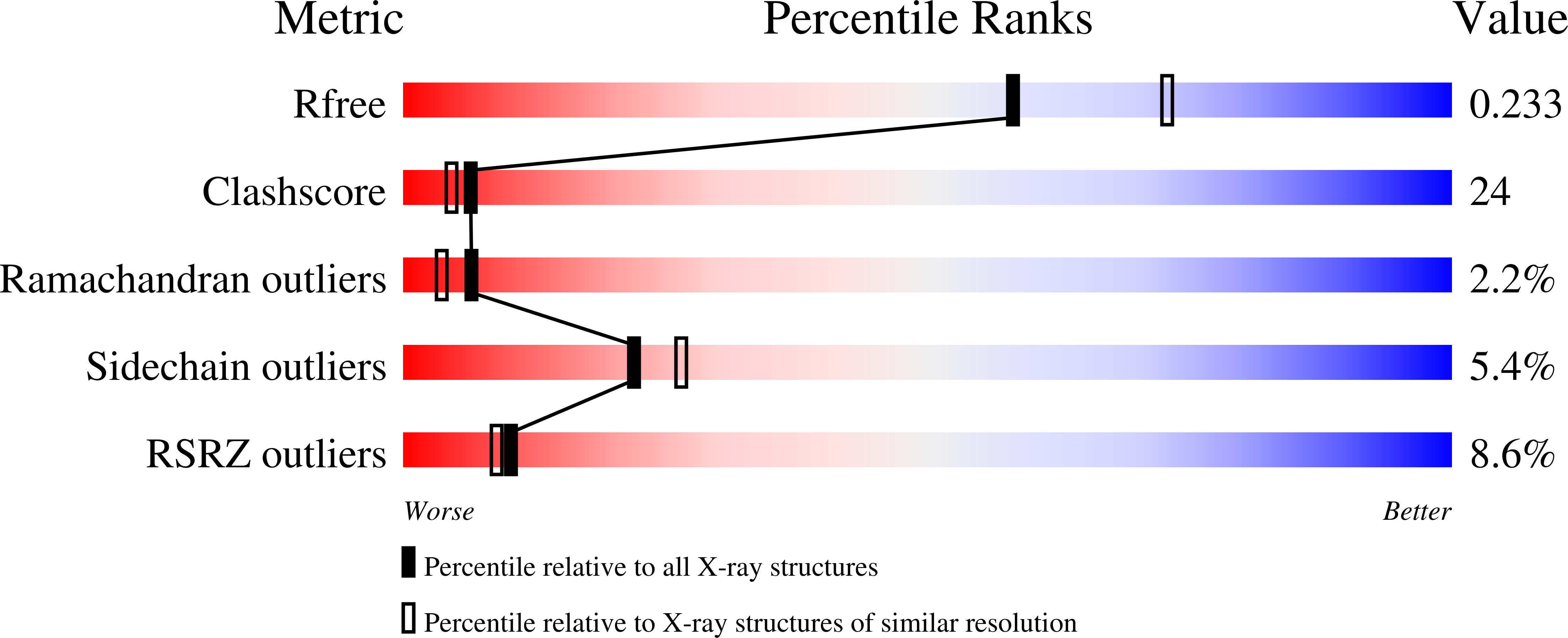
Bacterial cell division requires accurate selection of the middle of the cell, where the bacterial tubulin homologue FtsZ polymerizes into a ring structure. In Escherichia coli, site selection is dependent on MinC, MinD and MINE: MinC acts, with MinD, to inhibit division at sites other than the midcell by directly interacting with FTSZ: Here we report the crystal structure to 2.2 A of MinC from Thermotoga maritima. MinC consists of two domains separated by a short linker. The C-terminal domain is a right-handed beta-helix and is involved in dimer formation. The crystals contain two different MinC dimers, demonstrating flexibility in the linker region. The two-domain architecture and dimerization of MinC can be rationalized with a model of cell division inhibition. MinC does not act like SulA, which affects the GTPase activity of FtsZ, and the model can explain how MinC would select for the FtsZ polymer rather than the monomer.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 2QH, UK.