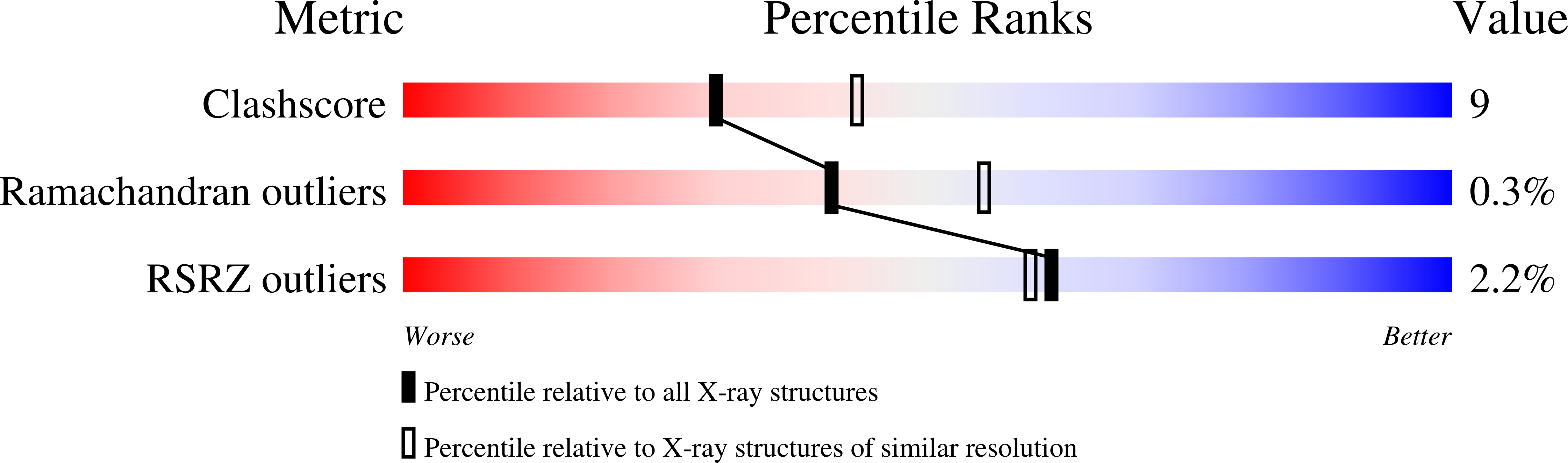
The Structure of an Alternative Form of Paracoccus Pantotrophus Cytochrome Cd1 Nitrite Reductase
Sjogren, T., Hajdu, J.(2001) J Biol Chem 276: 29450
- PubMed: 11373294
- DOI: https://doi.org/10.1074/jbc.M103657200
- Primary Citation of Related Structures:
1H9X, 1H9Y, 1HCM - PubMed Abstract:
Cytochrome cd(1) nitrite reductase is a bifunctional enzyme, which can catalyze the 1-electron reduction of nitrite to nitric oxide and the 4-electron reduction of dioxygen to water. Here we describe the structure of reduced nitrite reductase, crystallized under anaerobic conditions. The structure reveals substantial domain rearrangements with the c domain rotated by 60 degrees and shifted by approximately 20 A compared with previously known structures from crystals grown under oxidizing conditions. This alternative conformation gives rise to different electron transfer routes between the c and d(1) domains and points to the involvement of elements of very large structural changes in the function in this enzyme. In the present structure, the c heme has a His-69/Met-106 ligation, and this ligation does not change upon oxidation in the crystal. The d(1) heme is penta-coordinated, and the d(1) iron is displaced from the heme plane by 0.5 A toward the proximal ligand, His-200. After oxidation, the iron moves into the d(1) heme plane. A surprising finding is that although reduced nitrite reductase can be readily oxidized by dioxygen in the new crystal, it cannot turnover with its other substrate, nitrite. The results suggest that the rearrangement of the domains affects catalysis and substrate selectivity.
Organizational Affiliation:
Department of Biochemistry, Uppsala University, Box 576, S-751 23 Uppsala, Sweden.