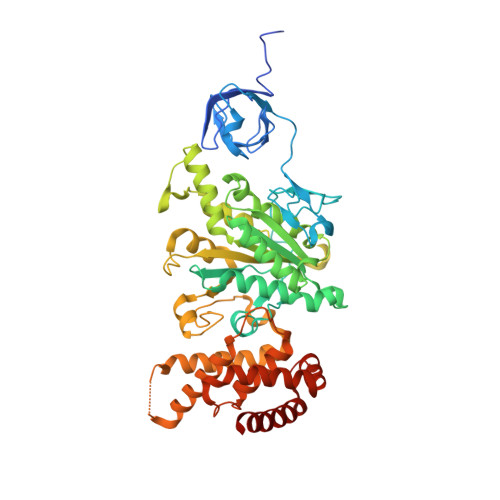
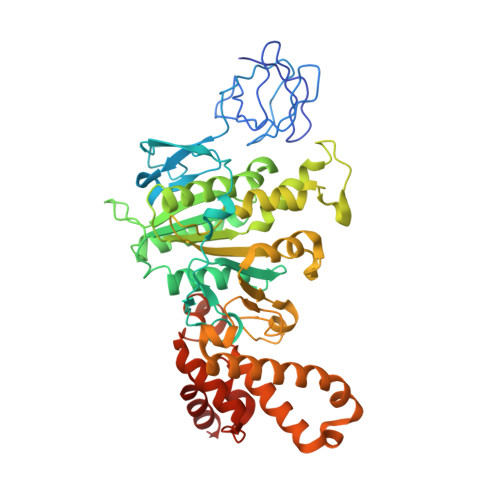
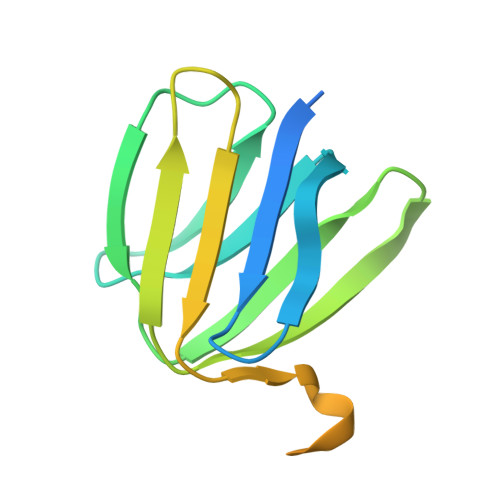
Structure of Bovine Mitochondrial F1-ATPase with Nucleotide Bound to All Three Catalytic Sites: Implications for the Mechanism of Rotary Catalysis
Menz, R.I., Walker, J.E., Leslie, A.G.W.(2001) Cell 106: 331
- PubMed: 11509182
- DOI: https://doi.org/10.1016/s0092-8674(01)00452-4
- Primary Citation of Related Structures:
1H8E - PubMed Abstract:
The crystal structure of a novel aluminium fluoride inhibited form of bovine mitochondrial F(1)-ATPase has been determined at 2 A resolution. In contrast to all previously determined structures of the bovine enzyme, all three catalytic sites are occupied by nucleotide. The subunit that did not bind nucleotide in previous structures binds ADP and sulfate (mimicking phosphate), and adopts a "half-closed" conformation. This structure probably represents the posthydrolysis, pre-product release step on the catalytic pathway. A catalytic scheme for hydrolysis (and synthesis) at physiological rates and a mechanism for the ATP-driven rotation of the gamma subunit are proposed based on the crystal structures of the bovine enzyme.
Organizational Affiliation:
Medical Research Council Laboratory of Molecular Biology, Hills Road, Cambridge CB2 2QH, United Kingdom.