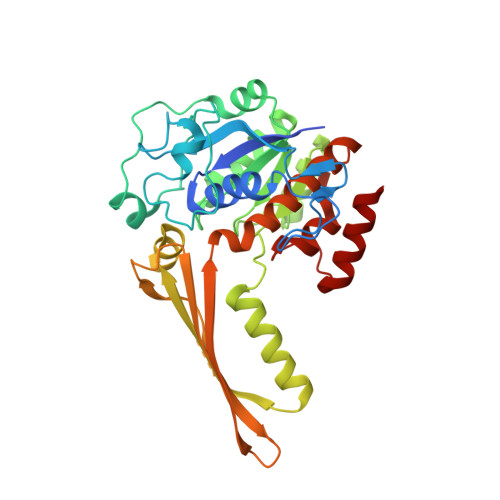
Crystal Structure of Inositol 1-Phosphate Synthase from Mycobacterium Tuberculosis, a Key Enzyme in Phosphatidylinositol Synthesis
Norman, R.A., Mcalister, M.S.B., Murray-Rust, J., Movahedzadeh, F., Stoker, N.G., Mcdonald, N.Q.(2002) Structure 10: 393
- PubMed: 12005437
- DOI: https://doi.org/10.1016/s0969-2126(02)00718-9
- Primary Citation of Related Structures:
1GR0 - PubMed Abstract:
Phosphatidylinositol (PI) is essential for Mycobacterium tuberculosis viability and the enzymes involved in the PI biosynthetic pathway are potential antimycobacterial agents for which little structural information is available. The rate-limiting step in the pathway is the production of (L)-myo-inositol 1-phosphate from (D)-glucose 6-phosphate, a complex reaction catalyzed by the enzyme inositol 1-phosphate synthase. We have determined the crystal structure of this enzyme from Mycobacterium tuberculosis (tbINO) at 1.95 A resolution, bound to the cofactor NAD+. The active site is located within a deep cleft at the junction between two domains. The unexpected presence of a zinc ion here suggests a mechanistic difference from the eukaryotic inositol synthases, which are stimulated by monovalent cations, that may be exploitable in developing selective inhibitors of tbINO.
Organizational Affiliation:
Structural Biology Laboratory, Cancer Research U K, London, United Kingdom.