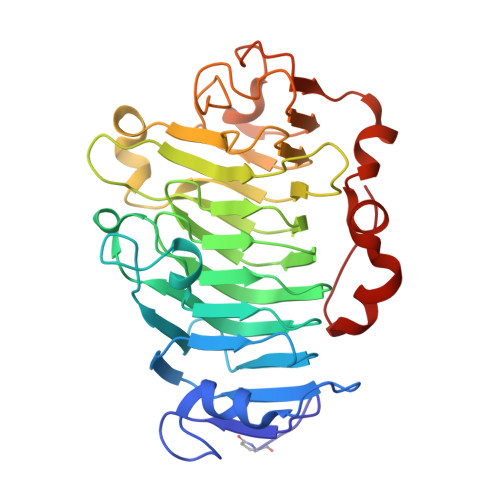
Crystal Structure of Plant Pectin Methylesterase
Johansson, K., El-Ahmad, M., Friemann, R., Jornvall, H., Markovic, O., Eklund, H.(2002) FEBS Lett 514: 243
- PubMed: 11943159
- DOI: https://doi.org/10.1016/s0014-5793(02)02372-4
- Primary Citation of Related Structures:
1GQ8 - PubMed Abstract:
Pectin is a principal component in the primary cell wall of plants. During cell development, pectin is modified by pectin methylesterases to give different properties to the cell wall. This report describes the first crystal structure of a plant pectin methylesterase. The beta-helical structure embodies a central cleft, lined by several aromatic residues, that has been deduced to be suitable for pectin binding. The active site is found at the center of this cleft where Asp157 is suggested to act as the nucleophile, Asp136 as an acid/base and Gln113/Gln135 to form an anion hole to stabilize the transition state.
Organizational Affiliation:
Department of Molecular Biology, Swedish University of Agricultural Sciences, S-751 24 Uppsala, Sweden.