Synthesis, Characterization and Solution Structure of Tethered Oligonucleotides Containing an Internal 3'-Phosphoglycolate, 5'-Phosphate Gapped Lesion
Junker, H.-D., Hoehn, S.T., Bunt, R.C., Marathius, V., Chen, J., Turner, C.J., Stubbe, J.(2002) Nucleic Acids Res 30: 5497-5508
- PubMed: 12490718
- DOI: https://doi.org/10.1093/nar/gkf681
- Primary Citation of Related Structures:
1G5K, 1GJ1, 1N0K, 1N0O - PubMed Abstract:
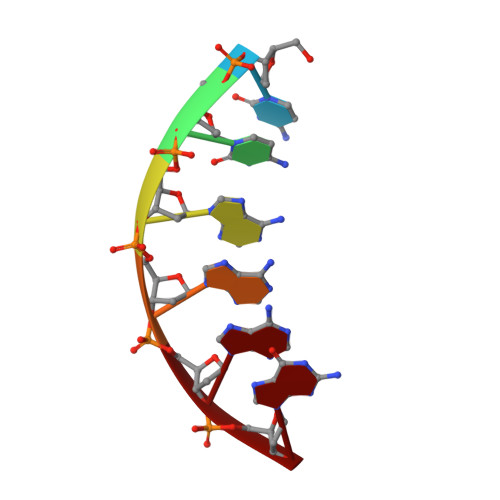
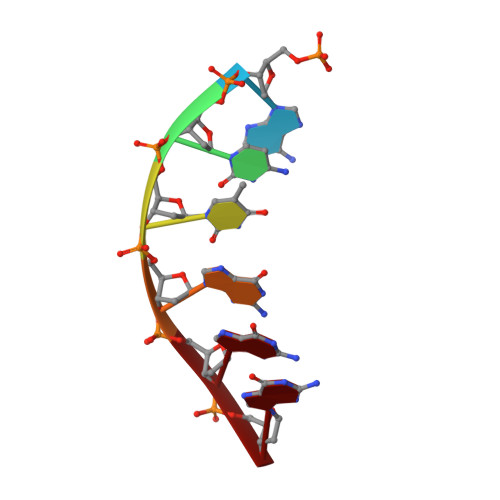
Bleomycins (BLMs) are antitumor antibiotics that in the presence of iron and oxygen mediate DNA damage by 4'-hydrogen atom abstraction of pyrimidines 3' to guanines. The resulting 4'-deoxyribose radicals can be trapped by O2 and ultimately result in the formation of base-propenal and gapped DNA with 3'-phosphoglycolate (3'-PG) and 5'-phosphate (5'-P) ends. The role of this lesion in triggering double-strand cleavage of duplex DNA by a single BLM molecule and the mechanism by which this lesion is repaired in vivo remain unsolved problems. The structure of these lesions is an essential step in addressing both of these problems. Duplex DNAs (13mers containing tethered hexaethylene glycol linkers) with GTAC and GGCC cleavage sites have been synthesized in which gaps containing 3'-PG and 5'-P ends at the sites of BLM cleavage have been inserted. The former sequence represents a hot spot for double-strand cleavage, while the latter is a hot spot for single-strand cleavage. Analytical methods to characterize the lesioned products have been developed. These oligonucleotides have been examined using 2D NMR methods and molecular modeling. The studies reveal that the lesioned DNAs are B-form and the 3'-PG and 5'-P are extrahelical. The base opposite the gap and the base pairs adjacent to the gap remain well stacked in the DNA duplex. Titrations of the lesioned GGCC oligomer with HOO-CoBLM leads to a mixture of complexes, in contrast to results of a similar titration with the lesioned GTAC oligomer.
Organizational Affiliation:
Department of Chemistry, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, USA.