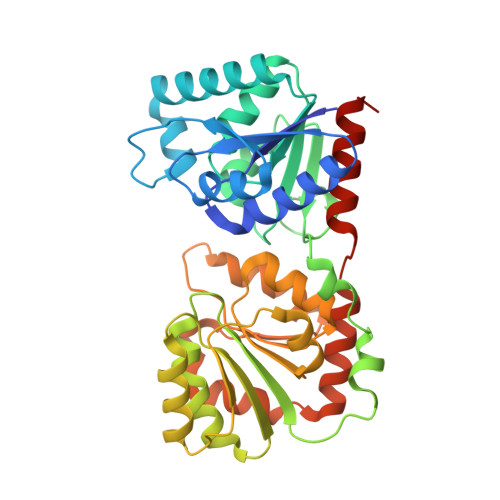
The 1.9 A crystal structure of Escherichia coli MurG, a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis.
Ha, S., Walker, D., Shi, Y., Walker, S.(2000) Protein Sci 9: 1045-1052
- PubMed: 10892798
- DOI: https://doi.org/10.1110/ps.9.6.1045
- Primary Citation of Related Structures:
1F0K - PubMed Abstract:
The 1.9 A X-ray structure of a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis is reported. This enzyme, MurG, contains two alpha/beta open sheet domains separated by a deep cleft. Structural analysis suggests that the C-terminal domain contains the UDP-GlcNAc binding site while the N-terminal domain contains the acceptor binding site and likely membrane association site. Combined with sequence data from other MurG homologs, this structure provides insight into the residues that are important in substrate binding and catalysis. We have also noted that a conserved region found in many UDP-sugar transferases maps to a beta/alpha/beta/alpha supersecondary structural motif in the donor binding region of MurG, an observation that may be helpful in glycosyltransferase structure prediction. The identification of a conserved structural motif involved in donor binding in different UDP-sugar transferases also suggests that it may be possible to identify--and perhaps alter--the residues that help determine donor specificity.
Organizational Affiliation:
Department of Chemistry, Princeton University, New Jersey 08544, USA.