Inhibition of EcoRV Endonuclease by Deoxyribo-3'-S-phosphorothiolates: A High-Resolution X-ray Crystallographic Study
Horton, N.C., Connolly, B.A., Perona, J.J.(2000) J Am Chem Soc 122: 3314-3324
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2000) J Am Chem Soc 122: 3314-3324
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| TYPE II RESTRICTION ENZYME ECORV | C [auth A], D [auth B] | 245 | Escherichia coli | Mutation(s): 0 EC: 3.1.21.4 | 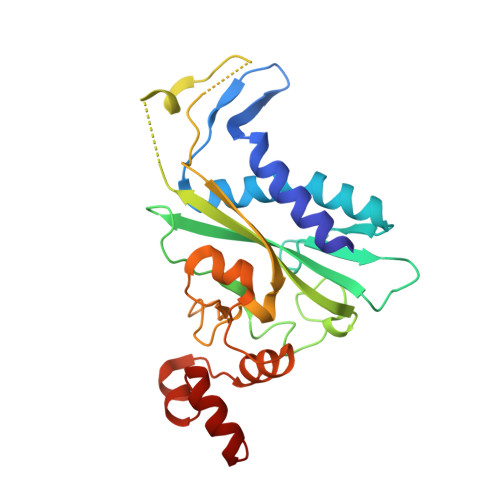 |
UniProt | |||||
Find proteins for P04390 (Escherichia coli) Explore P04390 Go to UniProtKB: P04390 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P04390 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*AP*AP*AP*GP*AP*(TSP)P*AP*TP*CP*TP*T)-3') | A [auth C] | 11 | N/A | 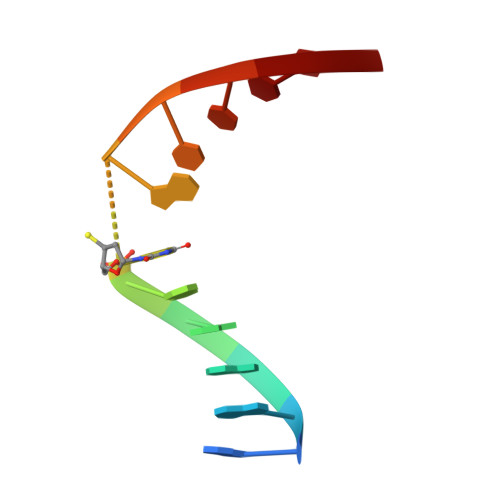 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*CP*AP*AP*GP*AP*(TSP)P*AP*TP*CP*TP*T)-3') | B [auth D] | 11 | N/A | 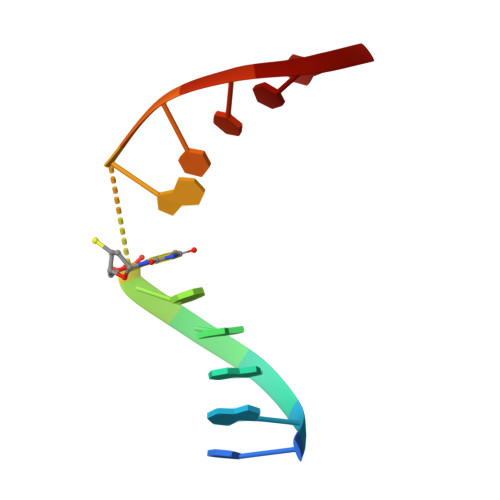 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ACY Query on ACY | G [auth A], J [auth B] | ACETIC ACID C2 H4 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-N |  | ||
| CL Query on CL | E [auth A], F [auth A], H [auth B], I [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 47.9 | α = 96.9 |
| b = 48.6 | β = 108.9 |
| c = 63.9 | γ = 106.8 |
| Software Name | Purpose |
|---|---|
| MOSFLM | data reduction |
| TRUNCATE | data reduction |
| X-PLOR | model building |
| X-PLOR | refinement |
| CCP4 | data scaling |
| X-PLOR | phasing |