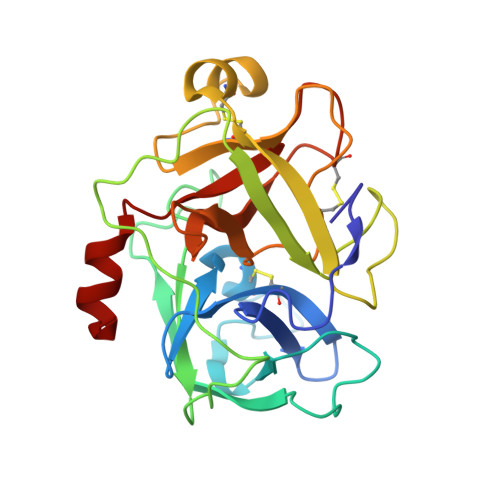
Catalytic Domain Structures of Mt-Sp1/Matriptase, a Matrix-Degrading Transmembrane Serine Proteinase.
Friedrich, R., Fuentes-Prior, P., Ong, E., Coombs, G., Hunter, M., Oehler, R., Pierson, D., Gonzalez, R., Huber, R., Bode, W., Madison, E.L.(2002) J Biol Chem 277: 2160
- PubMed: 11696548
- DOI: https://doi.org/10.1074/jbc.M109830200
- Primary Citation of Related Structures:
1EAW, 1EAX - PubMed Abstract:
The type II transmembrane multidomain serine proteinase MT-SP1/matriptase is highly expressed in many human cancer-derived cell lines and has been implicated in extracellular matrix re-modeling, tumor growth, and metastasis. We have expressed the catalytic domain of MT-SP1 and solved the crystal structures of complexes with benzamidine at 1.3 A and bovine pancreatic trypsin inhibitor at 2.9 A. MT-SP1 exhibits a trypsin-like serine proteinase fold, featuring a unique nine-residue 60-insertion loop that influences interactions with protein substrates. The structure discloses a trypsin-like S1 pocket, a small hydrophobic S2 subsite, and an open negatively charged S4 cavity that favors the binding of basic P3/P4 residues. A complementary charge pattern on the surface opposite the active site cleft suggests a distinct docking of the preceding low density lipoprotein receptor class A domain. The benzamidine crystals possess a freely accessible active site and are hence well suited for soaking small molecules, facilitating the improvement of inhibitors. The crystal structure of the MT-SP1 complex with bovine pancreatic trypsin inhibitor serves as a model for hepatocyte growth factor activator inhibitor 1, the physiological inhibitor of MT-SP1, and suggests determinants for the substrate specificity.
Organizational Affiliation:
Max-Planck-Institut für Biochemie, Abteilung Strukturforschung, Am Klopferspitz 18a, 82152 Martinsried, Germany.