The MAD1-Sin3B Interaction Involves a Novel Helical Fold
Spronk, C.A.E.M., Tessari, M., Kaan, A.M., Jansen, J.F.A., Vermeulen, M., Stunnenberg, H.G., Vuister, G.W.(2000) Nat Struct Biol 7: 1100-1104
- PubMed: 11101889
- DOI: https://doi.org/10.1038/81944
- Primary Citation of Related Structures:
1E91 - PubMed Abstract:
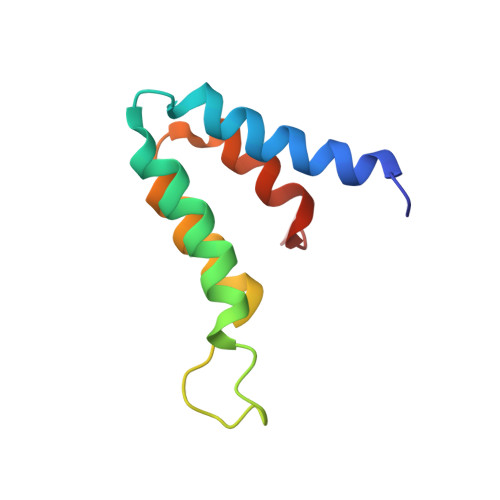
Sin3A or Sin3B are components of a corepressor complex that mediates repression by transcription factors such as the helix-loop-helix proteins Mad and Mxi. Members of the Mad/Mxi family of repressors play important roles in the transition between proliferation and differentiation by down-regulating the expression of genes that are activated by the proto-oncogene product Myc. Here, we report the solution structure of the second paired amphipathic helix (PAH) domain (PAH2) of Sin3B in complex with a peptide comprising the N-terminal region of Mad1. This complex exhibits a novel interaction fold for which we propose the name 'wedged helical bundle'. Four alpha-helices of PAH2 form a hydrophobic cleft that accommodates an amphipathic Mad1 alpha-helix. Our data further show that, upon binding Mad1, secondary structure elements of PAH2 are stabilized. The PAH2-Mad1 structure provides the basis for determining the principles of protein interaction and selectivity involving PAH domains.
Organizational Affiliation:
Department of Biophysical Chemistry, NSR Center, University of Nijmegen, The Netherlands.