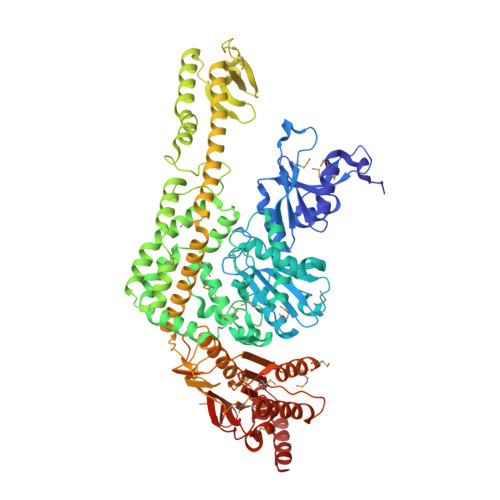
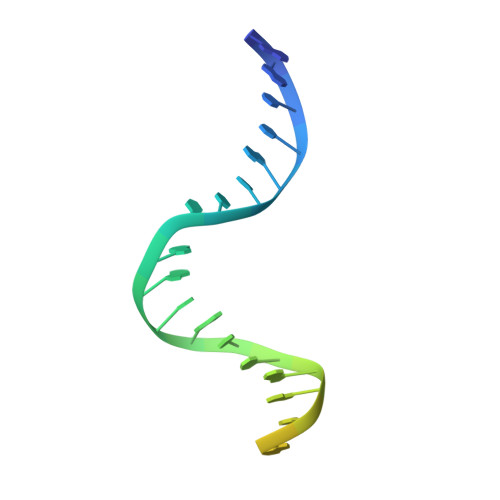
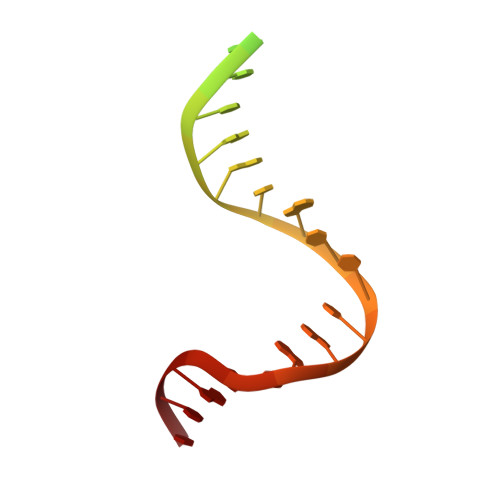
The Crystal Structure of DNA Mismatch Repair Protein Muts Binding to a G X T Mismatch
Lamers, M.H., Perrakis, A., Enzlin, J.H., Winterwerp, H.H.K., De Wind, N., Sixma, T.K.(2000) Nature 407: 711
- PubMed: 11048711
- DOI: https://doi.org/10.1038/35037523
- Primary Citation of Related Structures:
1E3M - PubMed Abstract:
DNA mismatch repair ensures genomic integrity on DNA replication. Recognition of a DNA mismatch by a dimeric MutS protein initiates a cascade of reactions and results in repair of the newly synthesized strand; however, details of the molecular mechanism remain controversial. Here we present the crystal structure at 2.2 A of MutS from Escherichia coli bound to a G x T mismatch. The two MutS monomers have different conformations and form a heterodimer at the structural level. Only one monomer recognizes the mismatch specifically and has ADP bound. Mismatch recognition occurs by extensive minor groove interactions causing unusual base pairing and kinking of the DNA. Nonspecific major groove DNA-binding domains from both monomers embrace the DNA in a clamp-like structure. The interleaved nucleotide-binding sites are located far from the DNA. Mutations in human MutS alpha (MSH2/MSH6) that lead to hereditary predisposition for cancer, such as hereditary non-polyposis colorectal cancer, can be mapped to this crystal structure.
Organizational Affiliation:
Division of Molecular Carcinogenesis, Netherlands Cancer Institute, Amsterdam.