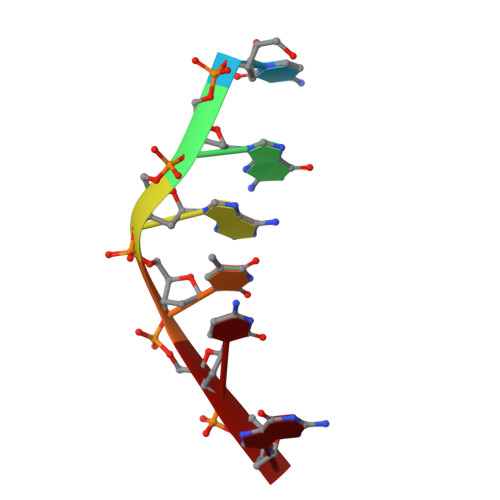
DNA-drug interactions. The crystal structure of d(CGATCG) complexed with daunomycin.
Moore, M.H., Hunter, W.N., d'Estaintot, B.L., Kennard, O.(1989) J Mol Biol 206: 693-705
- PubMed: 2738914
- DOI: https://doi.org/10.1016/0022-2836(89)90577-9
- Primary Citation of Related Structures:
1DA0 - PubMed Abstract:
The structure of a d(CGATCG)-daunomycin complex has been determined by single crystal X-ray diffraction techniques. Refinement, with the location of 40 solvent molecules, using data up to 1.5 A, converged with a final crystallographic residual, R = 0.25 (RW = 0.22). The tetragonal crystals are in space group P4(1)2(1)2, with cell dimensions of a = 27.98 A and c = 52.87 A. The self-complementary d(CGATCG) forms a distorted right-handed helix with a daunomycin molecule intercalated at each d(CpG) step. The daunomycin aglycon chromophore is oriented at right-angles to the long axis of the DNA base-pairs. This head-on intercalation is stabilized by direct hydrogen bonds and indirectly via solvent-mediated, hydrogen-bonding interactions between the chromophore and its intercalation site base-pairs. The cyclohexene ring and amino sugar substituent lie in the minor groove. The amino sugar N-3' forms a hydrogen bond with O-2 of the next neighbouring thymine. This electrostatic interaction helps position the sugar in a way that results in extensive van der Waals contacts between the drug and the DNA. There is no interaction between daunosamine and the DNA sugar-phosphate backbone. We present full experimental details and all relevant conformational parameters, and use the comparison with a d(CGTACG)-daunomycin complex to rationalize some neighbouring sequence effects involved in daunomycin binding.
Organizational Affiliation:
University Chemical Laboratory, Cambridge, U.K.