Crystal structure of gingipain R: an Arg-specific bacterial cysteine proteinase with a caspase-like fold.
Eichinger, A., Beisel, H.G., Jacob, U., Huber, R., Medrano, F.J., Banbula, A., Potempa, J., Travis, J., Bode, W.(1999) EMBO J 18: 5453-5462
- PubMed: 10523290
- DOI: https://doi.org/10.1093/emboj/18.20.5453
- Primary Citation of Related Structures:
1CVR - PubMed Abstract:
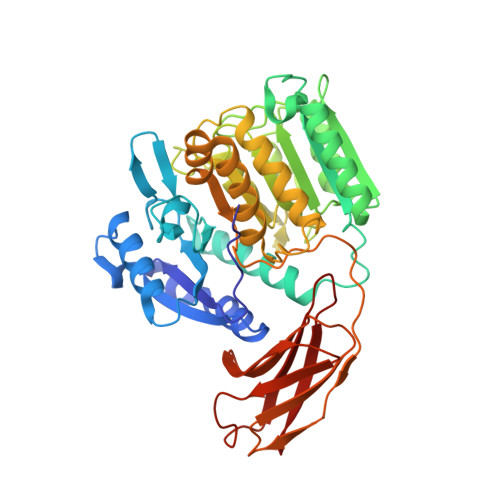
Gingipains are cysteine proteinases acting as key virulence factors of the bacterium Porphyromonas gingivalis, the major pathogen in periodontal disease. The 1.5 and 2.0 A crystal structures of free and D-Phe-Phe-Arg-chloromethylketone-inhibited gingipain R reveal a 435-residue, single-polypeptide chain organized into a catalytic and an immunoglobulin-like domain. The catalytic domain is subdivided into two subdomains comprising four- and six-stranded beta-sheets sandwiched by alpha-helices. Each subdomain bears topological similarities to the p20-p10 heterodimer of caspase-1. The second subdomain harbours the Cys-His catalytic diad and a nearby Glu arranged around the S1 specificity pocket, which carries an Asp residue to enforce preference for Arg-P1 residues. This gingipain R structure is an excellent template for the rational design of drugs with a potential to cure and prevent periodontitis. Here we show the binding mode of an arginine-containing inhibitor in the active-site, thus identifying major interaction sites defining a suitable pharmacophor.
Organizational Affiliation:
Max-Planck-Institut für Biochemie, Abteilung Strukturforschung, D-82152 Martinsried, Germany.