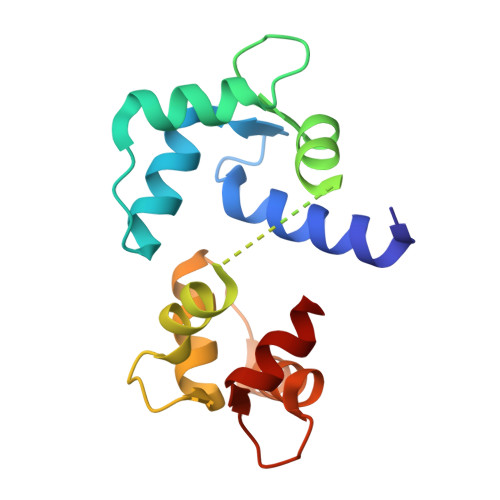
Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures.
Meador, W.E., Means, A.R., Quiocho, F.A.(1993) Science 262: 1718-1721
- PubMed: 8259515
- DOI: https://doi.org/10.1126/science.8259515
- Primary Citation of Related Structures:
1CDM - PubMed Abstract:
Calmodulin is the primary calcium-dependent signal transducer and regulator of a wide variety of essential cellular functions. The structure of calcium-calmodulin bound to the peptide corresponding to the calmodulin-binding domain of brain calmodulin-dependent protein kinase II alpha was determined to 2 angstrom resolution. A comparison to two other calcium-calmodulin structures reveals how the central helix unwinds in order to position the two domains optimally in the recognition of different target enzymes and clarifies the role of calcium in maintaining recognition-competent domain structures.
Organizational Affiliation:
Howard Hughes Medical Institute, Baylor College of Medicine, Houston, TX 77030.