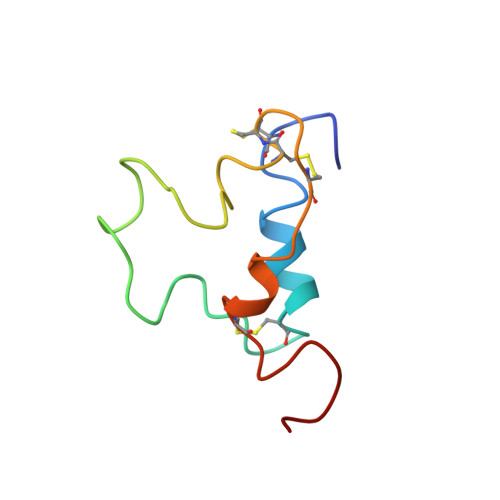
Three-dimensional structure of human insulin-like growth factor-I (IGF-I) determined by 1H-NMR and distance geometry.
Sato, A., Nishimura, S., Ohkubo, T., Kyogoku, Y., Koyama, S., Kobayashi, M., Yasuda, T., Kobayashi, Y.(1993) Int J Pept Protein Res 41: 433-440
- PubMed: 8391516
- DOI: https://doi.org/10.1111/j.1399-3011.1993.tb00462.x
- Primary Citation of Related Structures:
1BQT - PubMed Abstract:
The three-dimensional structure of human insulin-like growth factor-I has been determined through a combination of NMR measurements and distance geometry calculations. A total of 320 interatomic distance constraints, including 12 related to the disulfide bridges, were used in these calculations. The resulting structure is characterized by the presence of three helical rods corresponding to the sequence regions, Ala8-Cys18, Gly42-Cys48 and Leu54-Cys61. Furthermore, a turn structure and an extended structure exist in the Gly19-Gly22 and Phe23-Asn26 regions, respectively. Neglecting the N- and C-termini, with their expectedly high degree of mobility as well as a fluctuating C-domain, the r.m.s.d. value is 1.9 A for backbone atoms. Those of the three alpha-helical regions are 1.0, 0.9 and 0.8 A, respectively, 1.8 A being that for the total backbone atoms participating in the formation of these three helices, showing the good convergence of their spatial arrangements. The overall structure obtained here shows that the human IGF-I molecule folds into a spatial structure very similar to that of insulin in an aqueous solution.
Organizational Affiliation:
Institute for Protein Research, Osaka University, Japan.