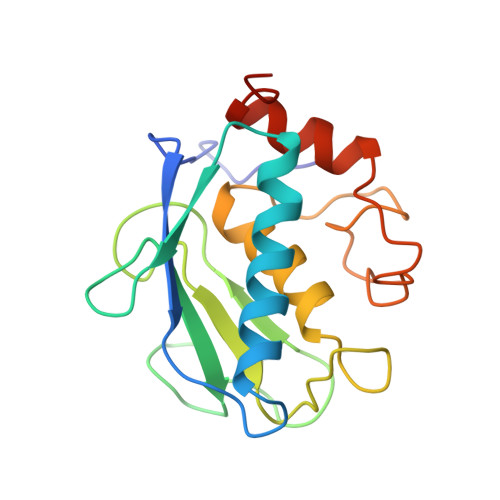
Crystal structure of the stromelysin catalytic domain at 2.0 A resolution: inhibitor-induced conformational changes.
Chen, L., Rydel, T.J., Gu, F., Dunaway, C.M., Pikul, S., Dunham, K.M., Barnett, B.L.(1999) J Mol Biol 293: 545-557
- PubMed: 10543949
- DOI: https://doi.org/10.1006/jmbi.1999.3147
- Primary Citation of Related Structures:
1B3D, 1CQR - PubMed Abstract:
Matrix metalloproteinases are believed to play an important role in pathological conditions such as osteoarthritis, rheumatoid arthritis and tumor invasion. Stromelysin is a zinc-dependent proteinase and a member of the matrix metalloproteinase family. We have solved the crystal structure of an active uninhibited form of truncated stromelysin and a complex with a hydroxamate-based inhibitor. The catalytic domain of the enzyme of residues 83-255 is an active fragment. Two crystallographically independent molecules, A and B, associate as a dimer in the crystals. There are three alpha-helices and one twisted, five-strand beta-sheet in each molecule, as well as one catalytic Zn, one structural Zn and three structural Ca ions. The active site of stromelysin is located in a large, hydrophobic cleft. In particular, the S1' specificity site is a deep and highly hydrophobic cavity. The structure of a hydroxamate-phosphinamide-type inhibitor-bound stromelysin complex, formed by diffusion soaking, has been solved as part of our structure-based design strategy. The most important feature we observed is an inhibitor-induced conformational change in the S1' cavity which is triggered by Tyr223. In the uninhibited enzyme structure, Tyr223 completely covers the S1' cavity, while in the complex, the P1' group of the inhibitor displaces the Tyr223 in order to fit into the S1' cavity. Furthermore, the displacement of Tyr223 induces a major conformational change of the entire loop from residue 222 to residue 231. This finding provides direct evidence that Tyr223 plays the role of gatekeeper of the S1' cavity. Another important intermolecular interaction occurs at the active sit of molecule A, in which the C-terminal tail (residues 251-255) from molecule B inserts. The C-terminal tail interacts extensively with the active site of molecule A, and the last residue (Thr255) coordinated to the catalytic zinc as the fourth ligand, much like a product inhibitor would. The inhibitor-induced conformational change and the intermolecular C-terminal-zinc coordination are significant in understanding the structure-activity relationships of the enzyme.
Organizational Affiliation:
The Procter & Gamble Company, Health Care Research Center, Mason, OH, 45040-9462, USA.