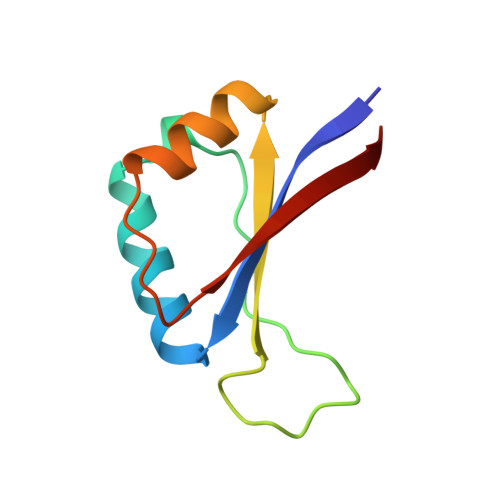
Structure of the E2 DNA-binding domain from human papillomavirus serotype 31 at 2.4 A.
Bussiere, D.E., Kong, X., Egan, D.A., Walter, K., Holzman, T.F., Lindh, F., Robins, T., Giranda, V.L.(1998) Acta Crystallogr D Biol Crystallogr 54: 1367-1376
- PubMed: 10089498
- DOI: https://doi.org/10.1107/s0907444998005587
- Primary Citation of Related Structures:
1A7G - PubMed Abstract:
The papillomaviruses are a family of small double-stranded DNA viruses which exclusively infect epithelial cells and stimulate the proliferation of those cells. A key protein within the papillomavirus life-cycle is known as the E2 (Early 2) protein and is responsible for regulating viral transcription from all viral promoters as well as for replication of the papillomavirus genome in tandem with another protein known as E1. The E2 protein itself consists of three functional domains: an N-terminal trans-activation domain, a proline-rich linker, and a C-terminal DNA-binding domain. The first crystal structure of the human papillomavirus, serotype 31 (HPV-31), E2 DNA-binding domain has been determined at 2.4 A resolution. The HPV DNA-binding domain monomer consists of two beta-alpha-beta repeats of approximately equal length and is arranged as to have an anti-parallel beta-sheet flanked by the two alpha-helices. The monomers form the functional in vivo dimer by association of the beta-sheets of each monomer so as to form an eight-stranded anti-parallel beta-barrel at the center of the dimer, with the alpha-helices lining the outside of the barrel. The overall structure of HVP-31 E2 DNA-binding domain is similar to both the bovine papillomavirus E2-binding domain and the Epstein-Barr nuclear antigen-1 DNA-binding domain.
Organizational Affiliation:
Division of Scientific Information, Analysis, and Management, Pharmaceutical Products Division, Abbott Laboratories, 100 Abbott Park Road, Abbott Park, IL 60064, USA.