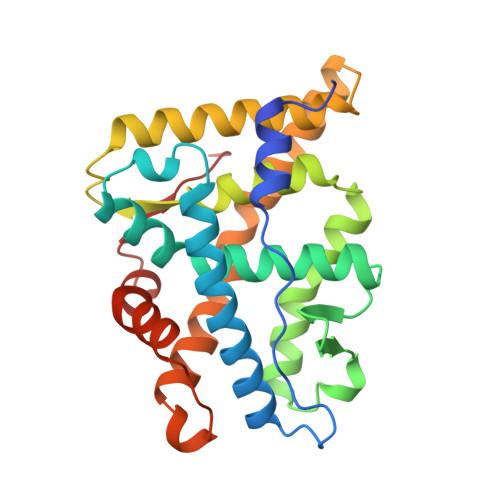
Atomic structure of progesterone complexed with its receptor.
Williams, S.P., Sigler, P.B.(1998) Nature 393: 392-396
- PubMed: 9620806
- DOI: https://doi.org/10.1038/30775
- Primary Citation of Related Structures:
1A28 - PubMed Abstract:
The physiological effects of progestins are mediated by the progesterone receptor, a member of the steroid/nuclear receptor superfamily. As progesterone is required for maintenance of pregnancy, its receptor has been a target for pharmaceuticals. Here we report the 1.8 A crystal structure of a progesterone-bound ligand-binding domain of the human progesterone receptor. The nature of this structure explains the receptor's selective affinity for progestins and establishes a common mode of recognition of 3-oxy steroids by the cognate receptors. Although the overall fold of the progesterone receptor is similar to that found in related receptors, the progesterone receptor has a quite different mode of dimerization. A hormone-induced stabilization of the carboxy-terminal secondary structure of the ligand-binding domain of the progesterone receptor accounts for the stereochemistry of this distinctive dimer, explains the receptor's characteristic pattern of ligand-dependent protease resistance and its loss of repression, and indicates how the anti-progestin RU486 might work in birth control. The structure also indicates that the analogous 3-keto-steroid receptors may have a similar mechanism of action.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, and the Howard Hughes Medical Institute, Yale University, New Haven, Connecticut 06510, USA.