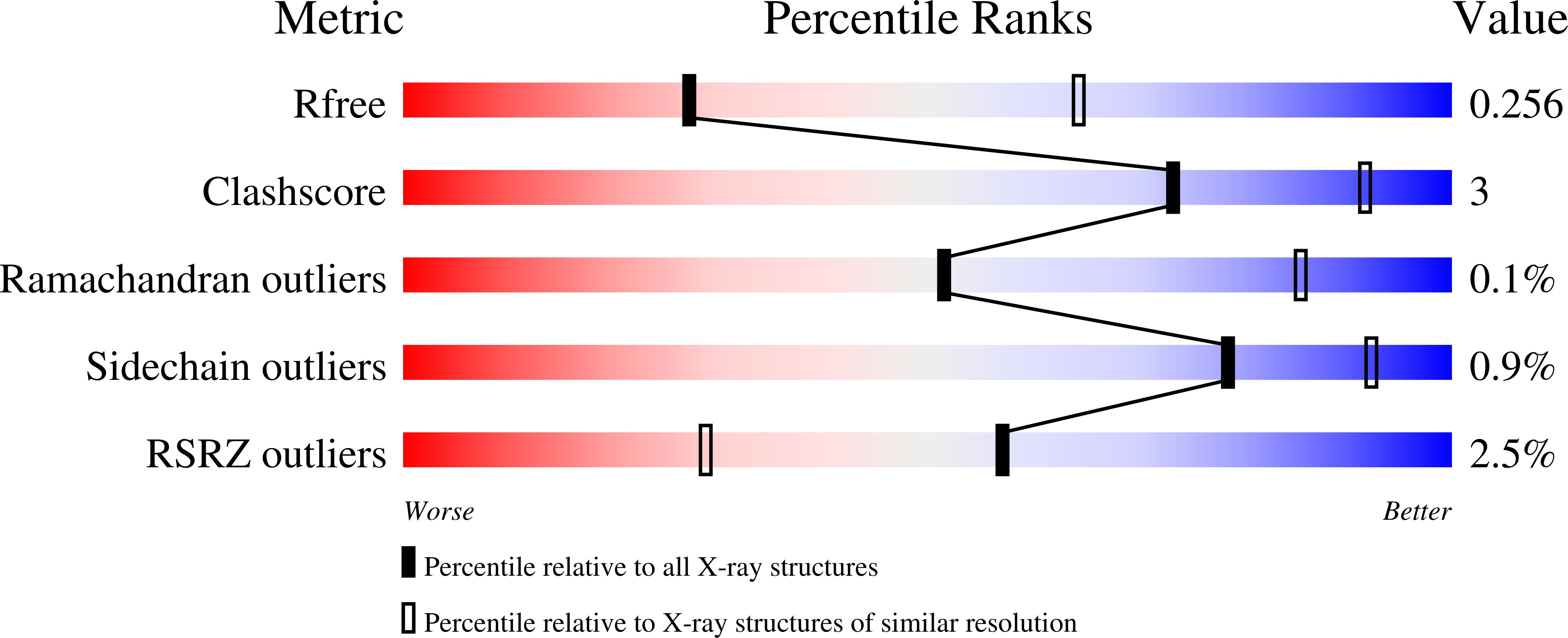
A conformation-selective monoclonal antibody against a small molecule-stabilised signalling-deficient form of TNF
Lightwood, D.J., Munro, R.J., Porter, J., McMillan, D., Carrington, B., Turner, A., Scott-Tucker, A., Hickford, E.S., Schmidt, A., Fox III, D., Maloney, A., Ceska, T., Bourne, T., O'Connell, J., Lawson, A.D.G.(2021) Nat Commun 12: 583-583
- PubMed: 33495445
- DOI: https://doi.org/10.1038/s41467-020-20825-6
- Primary Citation of Related Structures:
7KPA, 7KPB - PubMed Abstract:
We have recently described the development of a series of small-molecule inhibitors of human tumour necrosis factor (TNF) that stabilise an open, asymmetric, signalling-deficient form of the soluble TNF trimer. Here, we describe the generation, characterisation, and utility of a monoclonal antibody that selectively binds with high affinity to the asymmetric TNF trimer-small molecule complex. The antibody helps to define the molecular dynamics of the apo TNF trimer, reveals the mode of action and specificity of the small molecule inhibitors, acts as a chaperone in solving the human TNF-TNFR1 complex crystal structure, and facilitates the measurement of small molecule target occupancy in complex biological samples. We believe this work defines a role for monoclonal antibodies as tools to facilitate the discovery and development of small-molecule inhibitors of protein-protein interactions.
Organizational Affiliation:
UCB Pharma, 208 Bath Road, Slough, SL1 3WE, UK. Daniel.Lightwood@ucb.com.