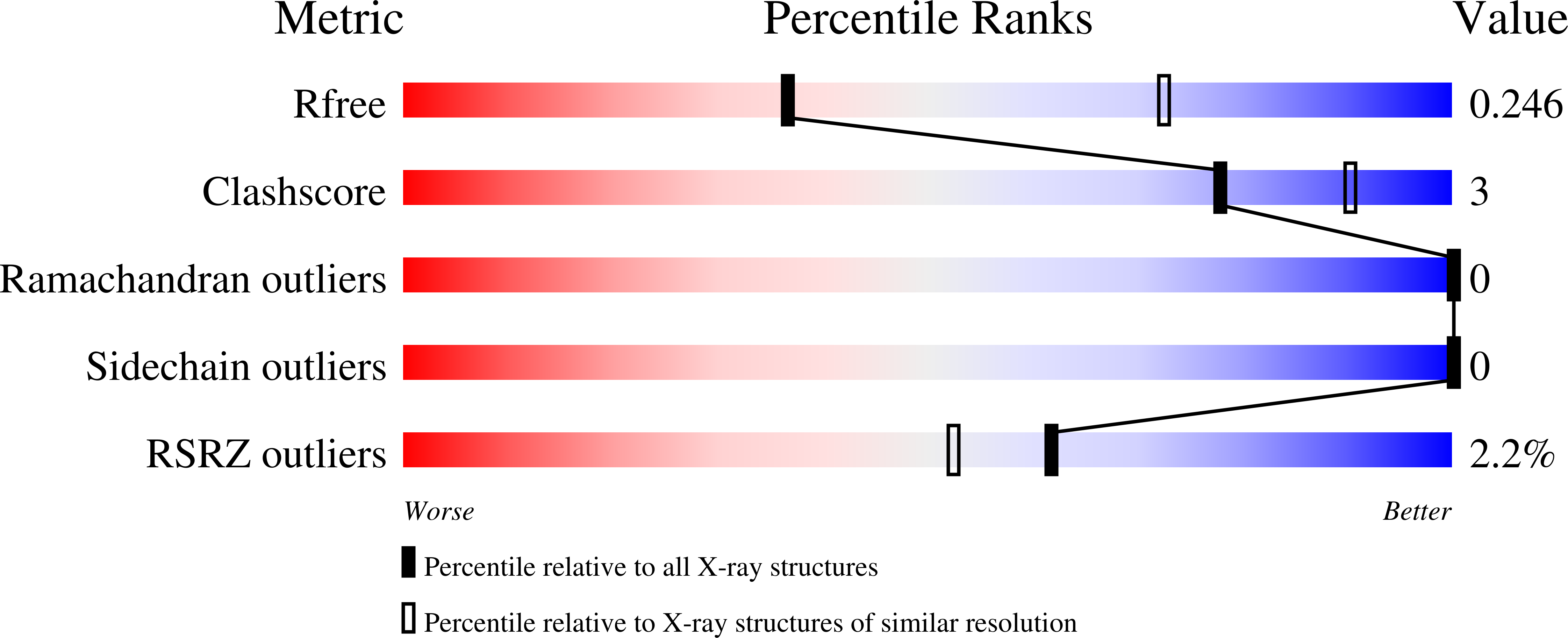
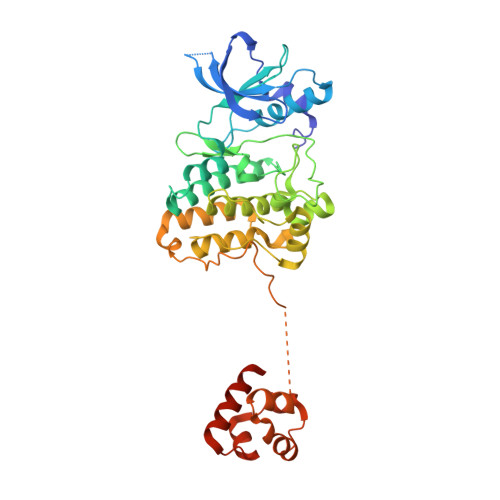
Regulation of the EphA2 receptor intracellular region by phosphomimetic negative charges in the kinase-SAM linker.
Lechtenberg, B.C., Gehring, M.P., Light, T.P., Horne, C.R., Matsumoto, M.W., Hristova, K., Pasquale, E.B.(2021) Nat Commun 12: 7047-7047
- PubMed: 34857764
- DOI: https://doi.org/10.1038/s41467-021-27343-z
- Primary Citation of Related Structures:
7KJA, 7KJB, 7KJC - PubMed Abstract:
Eph receptor tyrosine kinases play a key role in cell-cell communication. Lack of structural information on the entire multi-domain intracellular region of any Eph receptor has hindered understanding of their signaling mechanisms. Here, we use integrative structural biology to investigate the structure and dynamics of the EphA2 intracellular region. EphA2 promotes cancer malignancy through a poorly understood non-canonical form of signaling involving serine/threonine phosphorylation of the linker connecting its kinase and SAM domains. We show that accumulation of multiple linker negative charges, mimicking phosphorylation, induces cooperative changes in the EphA2 intracellular region from more closed to more extended conformations and perturbs the EphA2 juxtamembrane segment and kinase domain. In cells, linker negative charges promote EphA2 oligomerization. We also identify multiple kinases catalyzing linker phosphorylation. Our findings suggest multiple effects of linker phosphorylation on EphA2 signaling and imply that coordination of different kinases is necessary to promote EphA2 non-canonical signaling.
Organizational Affiliation:
Cancer Center, Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA, 92037, USA. lechtenberg.b@wehi.edu.au.