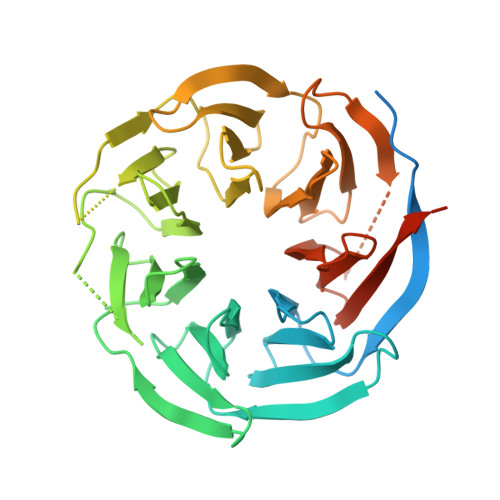
Crystal structure of the WD40 domain of human PLRG1.
Wang, X., Li, Y., Dai, H., Xu, C.(2021) Biochem Biophys Res Commun 534: 474-477
- PubMed: 33239170
- DOI: https://doi.org/10.1016/j.bbrc.2020.11.057
- Primary Citation of Related Structures:
7DH6 - PubMed Abstract:
PLRG1 is a evolutionarily conserved protein in spliceosome and plays an important role in maintaining the integral part of the splicoeosme and its proper splicing. Here we solved the high resolution crystal structure of the WD40 domain of human PLRG1 by crystallography and compared our crystal structure with the cryo-EM structure of PLRG1 bound with other splicing factors. We found that two loops of the WD40 domain become resolved upon binding to the proteins within the spliceosome. Thus our work characterize the dynamic property of PLRG1 during the spliceosome assembly by presenting its apo structure.
Organizational Affiliation:
MOE Key Laboratory for Membraneless Organelles and Cellular Dynamics, Hefei National Laboratory for Physical Sciences at the Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, 230027, China.