Cellular Synthesis and X-ray Crystal Structure of a Designed Protein Heterocatenane.
Liu, Y., Duan, Z., Fang, J., Zhang, F., Xiao, J., Zhang, W.B.(2020) Angew Chem Int Ed Engl 59: 16122-16127
- PubMed: 32506656
- DOI: https://doi.org/10.1002/anie.202005490
- Primary Citation of Related Structures:
7BWN - PubMed Abstract:
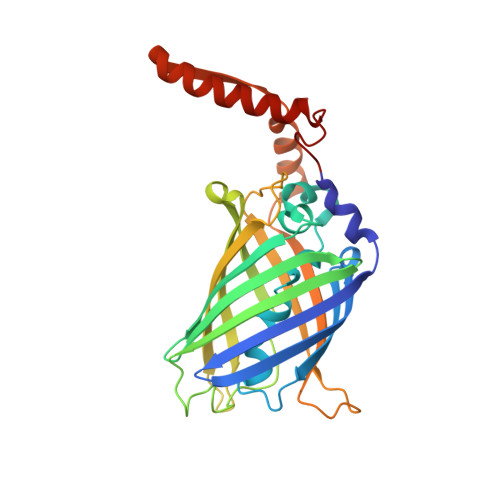
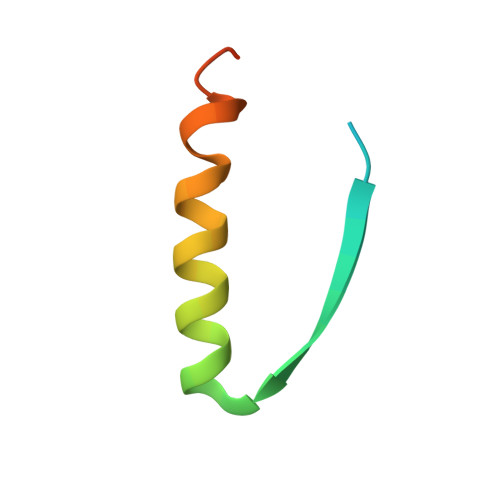
Herein, we report the biosynthesis of protein heterocatenanes using a programmed sequence of multiple post-translational processing events including intramolecular chain entanglement, in situ backbone cleavage, and spontaneous cyclization. The approach is general, autonomous, and can obviate the need for any additional enzymes. The catenane topology was convincingly proven using a combination of SDS-PAGE, LC-MS, size exclusion chromatography, controlled proteolytic digestion, and protein crystallography. The X-ray crystal structure clearly shows two mechanically interlocked protein rings with intact folded domains. It opens new avenues in the nascent field of protein-topology engineering.
Organizational Affiliation:
Beijing National Laboratory for Molecular Sciences, Key Laboratory of Polymer Chemistry &, Physics of Ministry of Education, Center for Soft Matter Science and Engineering, College of Chemistry and Molecular Engineering, Peking University, Beijing, 100871, P. R. China.