Anticancer immunity induced by a synthetic tumor-targeted CD137 agonist.
Upadhyaya, P., Lahdenranta, J., Hurov, K., Battula, S., Dods, R., Haines, E., Kleyman, M., Kristensson, J., Kublin, J., Lani, R., Ma, J., Mudd, G., Repash, E., Van Rietschoten, K., Stephen, T., You, F., Harrison, H., Chen, L., McDonnell, K., Brandish, P., Keen, N.(2021) J Immunother Cancer 9
- PubMed: 33500260
- DOI: https://doi.org/10.1136/jitc-2020-001762
- Primary Citation of Related Structures:
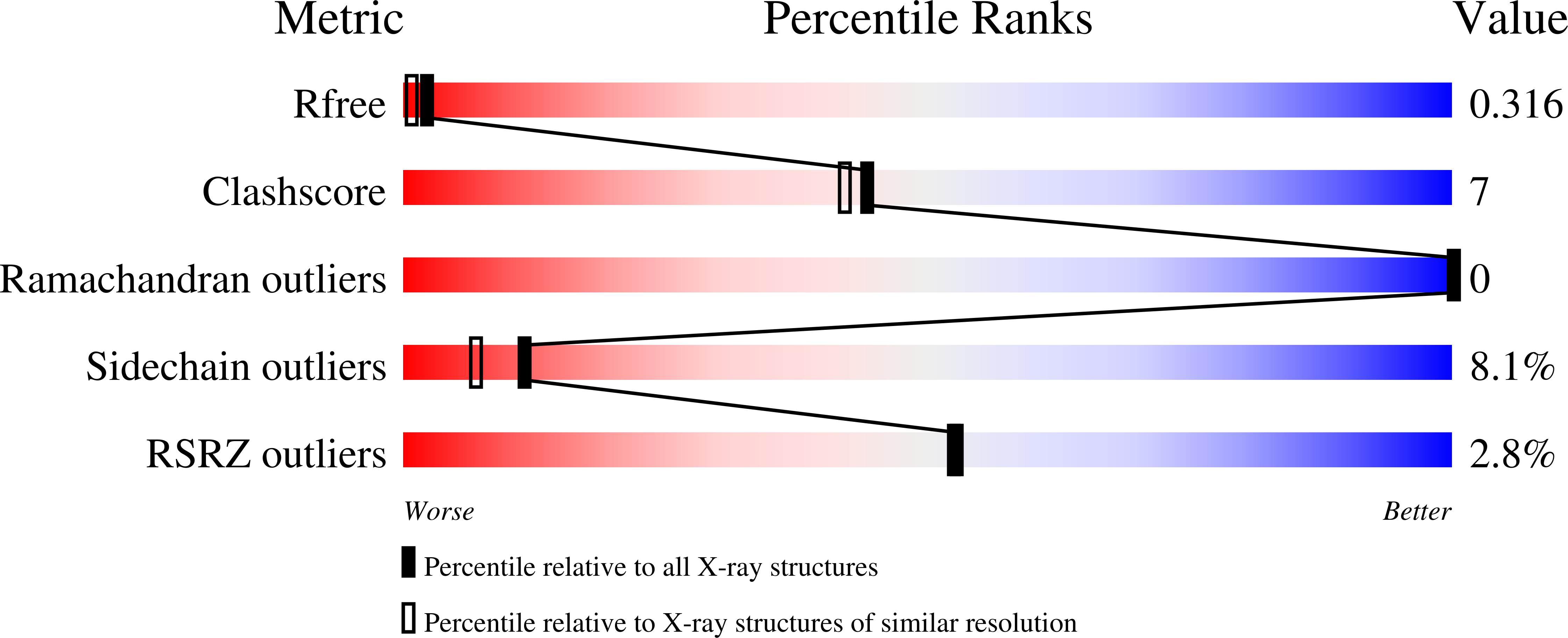
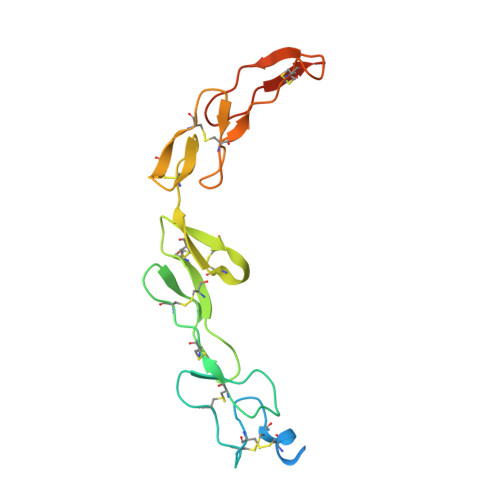
6Y8K - PubMed Abstract:
In contrast to immune checkpoint inhibitors, the use of antibodies as agonists of immune costimulatory receptors as cancer therapeutics has largely failed. We sought to address this problem using a new class of modular synthetic drugs, termed tumor-targeted immune cell agonists (TICAs), based on constrained bicyclic peptides ( Bicycles ).
Organizational Affiliation:
Bicycle Therapeutics, Lexington, Massachusetts, USA.