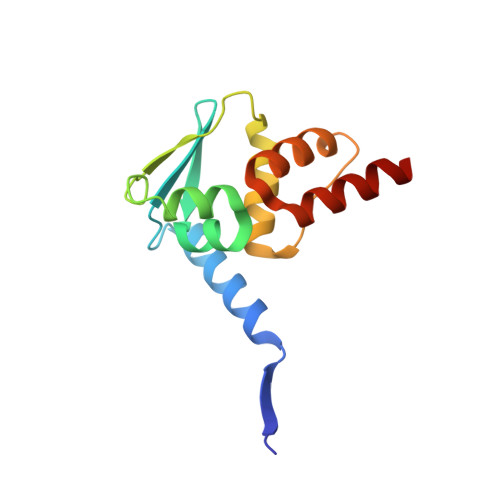
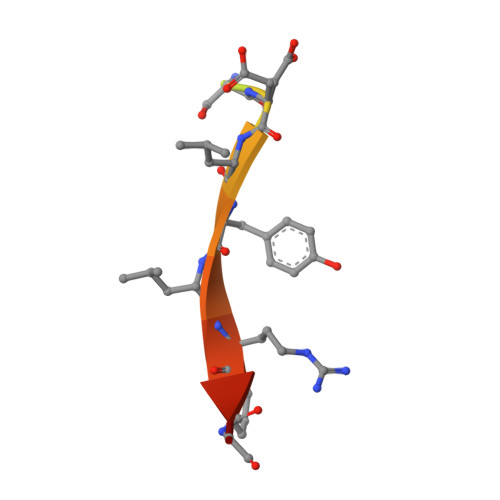
Functionalization of the BCL6 BTB domain into a noncovalent crystallization chaperone.
Zacharchenko, T., Wright, S.(2021) IUCrJ 8: 154-160
- PubMed: 33708392
- DOI: https://doi.org/10.1107/S2052252520015754
- Primary Citation of Related Structures:
6XWF, 6XXS, 6XYX, 6XZZ, 6Y17, 6ZBU - PubMed Abstract:
The production of diffraction-quality protein crystals is challenging and often requires bespoke, time-consuming and expensive strategies. A system has been developed in which the BCL6 BTB domain acts as a crystallization chaperone and promiscuous assembly block that may form the basis for affinity-capture crystallography. The protein of interest is expressed with a C-terminal tag that interacts with the BTB domain, and co-crystallization leads to its incorporation within a BTB-domain lattice. This strategy was used to solve the structure of the SH3 domain of human nebulin, a structure previously solved by NMR, at 1.56 Å resolution. This approach is simple and effective, requiring only routine protein complexation and crystallization screening, and should be applicable to a range of proteins.
Organizational Affiliation:
School of Biology and Astbury Centre for Structural Molecular Biology, University of Leeds, Woodhouse, Leeds LS2 9JT, United Kingdom.