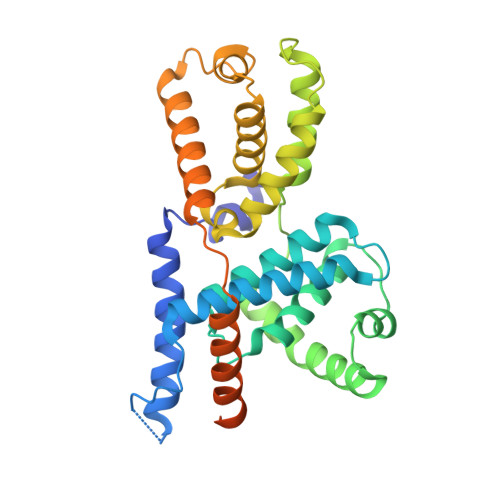
The cryoelectron microscopy structure of the human CDK-activating kinase.
Greber, B.J., Perez-Bertoldi, J.M., Lim, K., Iavarone, A.T., Toso, D.B., Nogales, E.(2020) Proc Natl Acad Sci U S A 117: 22849-22857
- PubMed: 32855301
- DOI: https://doi.org/10.1073/pnas.2009627117
- Primary Citation of Related Structures:
6XBZ, 6XD3 - PubMed Abstract:
The human CDK-activating kinase (CAK), a complex composed of cyclin-dependent kinase (CDK) 7, cyclin H, and MAT1, is a critical regulator of transcription initiation and the cell cycle. It acts by phosphorylating the C-terminal heptapeptide repeat domain of the RNA polymerase II (Pol II) subunit RPB1, which is an important regulatory event in transcription initiation by Pol II, and it phosphorylates the regulatory T-loop of CDKs that control cell cycle progression. Here, we have determined the three-dimensional (3D) structure of the catalytic module of human CAK, revealing the structural basis of its assembly and providing insight into CDK7 activation in this context. The unique third component of the complex, MAT1, substantially extends the interaction interface between CDK7 and cyclin H, explaining its role as a CAK assembly factor, and it forms interactions with the CDK7 T-loop, which may contribute to enhancing CAK activity. We have also determined the structure of the CAK in complex with the covalently bound inhibitor THZ1 in order to provide insight into the binding of inhibitors at the CDK7 active site and to aid in the rational design of therapeutic compounds.
Organizational Affiliation:
California Institute for Quantitative Biosciences (QB3), University of California, Berkeley, CA 94720; basil.greber@icr.ac.uk ENogales@lbl.gov.