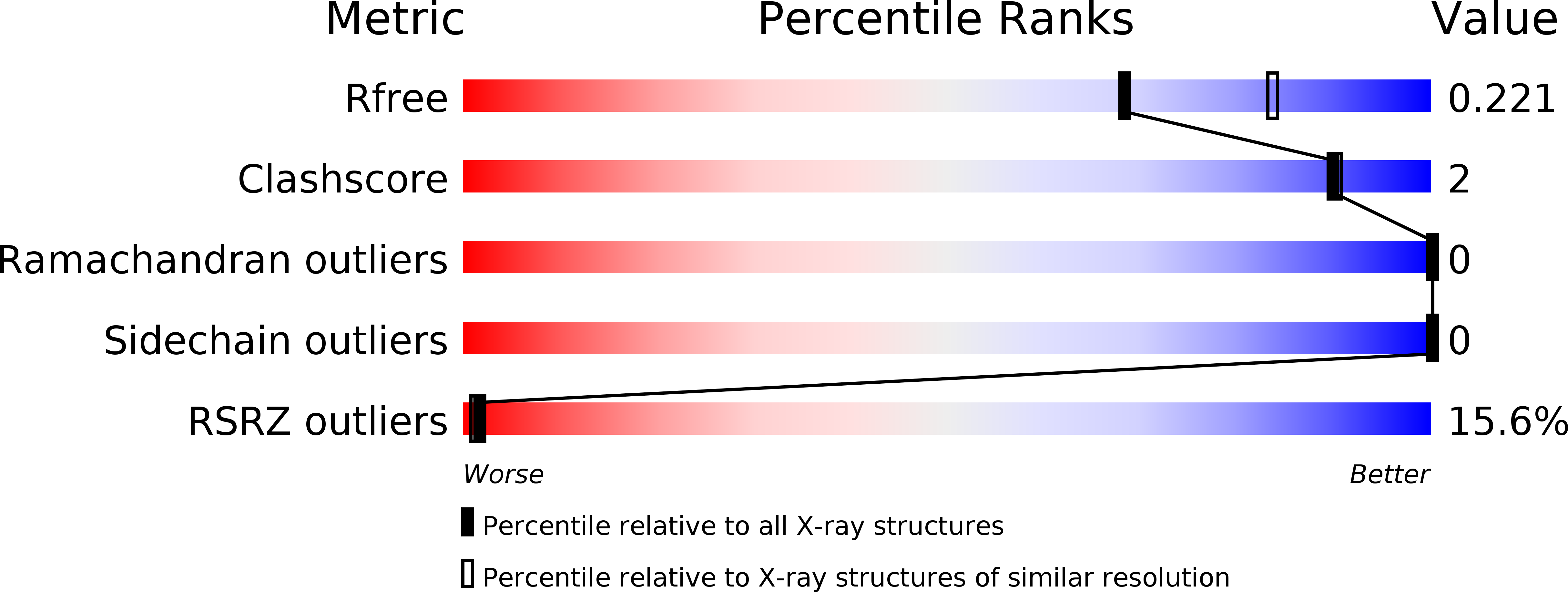
Discovery of hydroxy pyrimidine Factor IXa inhibitors.
Jayne, C.L., Andreani, T., Chan, T.Y., Chelliah, M.V., Clasby, M.C., Dwyer, M., Eagen, K.A., Fried, S., Greenlee, W.J., Guo, Z., Hawes, B., Hruza, A., Ingram, R., Keertikar, K.M., Neelamkavil, S., Reichert, P., Xia, Y., Chackalamannil, S.(2020) Bioorg Med Chem Lett 30: 127279-127279
- PubMed: 32527459
- DOI: https://doi.org/10.1016/j.bmcl.2020.127279
- Primary Citation of Related Structures:
6X5J, 6X5L, 6X5P - PubMed Abstract:
The synthesis and structure activity relationship development of a pyrimidine series of heterocyclic Factor IXa inhibitors is described. Increased selectivity over Factor Xa inhibition was achieved through SAR expansion of the P1 element. Select compounds were evaluated in vivo to assess their plasma levels in rat.
Organizational Affiliation:
Department of Discovery Chemistry, Merck & Co., Inc., Kenilworth, NJ 07033, USA. Electronic address: Charles.Jayne@merck.com.