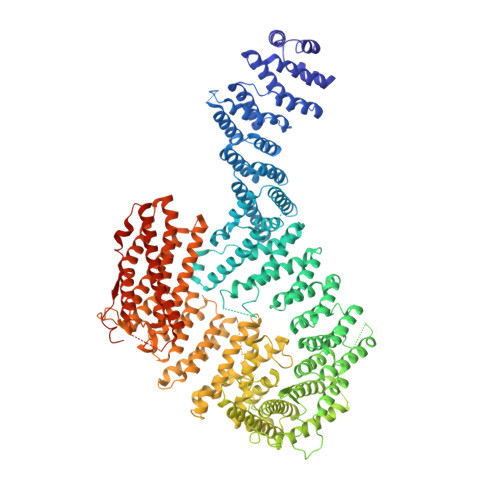
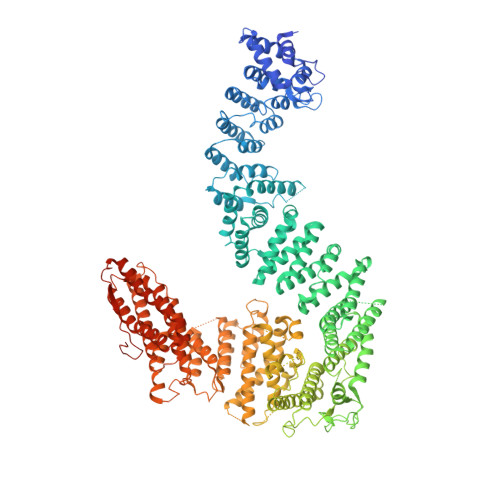
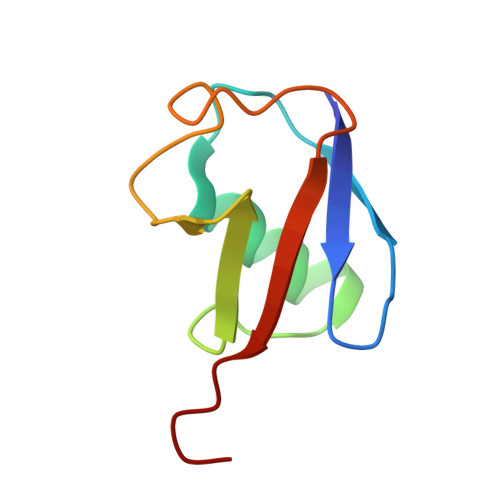
DNA clamp function of the monoubiquitinated Fanconi anaemia ID complex.
Wang, R., Wang, S., Dhar, A., Peralta, C., Pavletich, N.P.(2020) Nature 580: 278-282
- PubMed: 32269332
- DOI: https://doi.org/10.1038/s41586-020-2110-6
- Primary Citation of Related Structures:
6VAA, 6VAD, 6VAE, 6VAF - PubMed Abstract:
The ID complex, involving the proteins FANCI and FANCD2, is required for the repair of DNA interstrand crosslinks (ICL) and related lesions 1 . These proteins are mutated in Fanconi anaemia, a disease in which patients are predisposed to cancer. The Fanconi anaemia pathway of ICL repair is activated when a replication fork stalls at an ICL 2 ; this triggers monoubiquitination of the ID complex, in which one ubiquitin molecule is conjugated to each of FANCI and FANCD2. Monoubiquitination of ID is essential for ICL repair by excision, translesion synthesis and homologous recombination; however, its function remains unknown 1,3 . Here we report a cryo-electron microscopy structure of the monoubiquitinated human ID complex bound to DNA, and reveal that it forms a closed ring that encircles the DNA. By comparison with the structure of the non-ubiquitinated ID complex bound to ICL DNA-which we also report here-we show that monoubiquitination triggers a complete rearrangement of the open, trough-like ID structure through the ubiquitin of one protomer binding to the other protomer in a reciprocal fashion. These structures-together with biochemical data-indicate that the monoubiquitinated ID complex loses its preference for ICL and related branched DNA structures, and becomes a sliding DNA clamp that can coordinate the subsequent repair reactions. Our findings also reveal how monoubiquitination in general can induce an alternative protein structure with a new function.
Organizational Affiliation:
Structural Biology Program, Memorial Sloan-Kettering Cancer Center, New York, NY, USA.