Early Drug-Discovery Efforts towards the Identification of EP300/CBP Histone Acetyltransferase (HAT) Inhibitors.
Huhn, A.J., Gardberg, A.S., Poy, F., Brucelle, F., Vivat, V., Cantone, N., Patel, G., Patel, C., Cummings, R., Sims, R., Levell, J., Audia, J.E., Bommi-Reddy, A., Wilson, J.E.(2020) ChemMedChem 15: 955-960
- PubMed: 32181984
- DOI: https://doi.org/10.1002/cmdc.202000007
- Primary Citation of Related Structures:
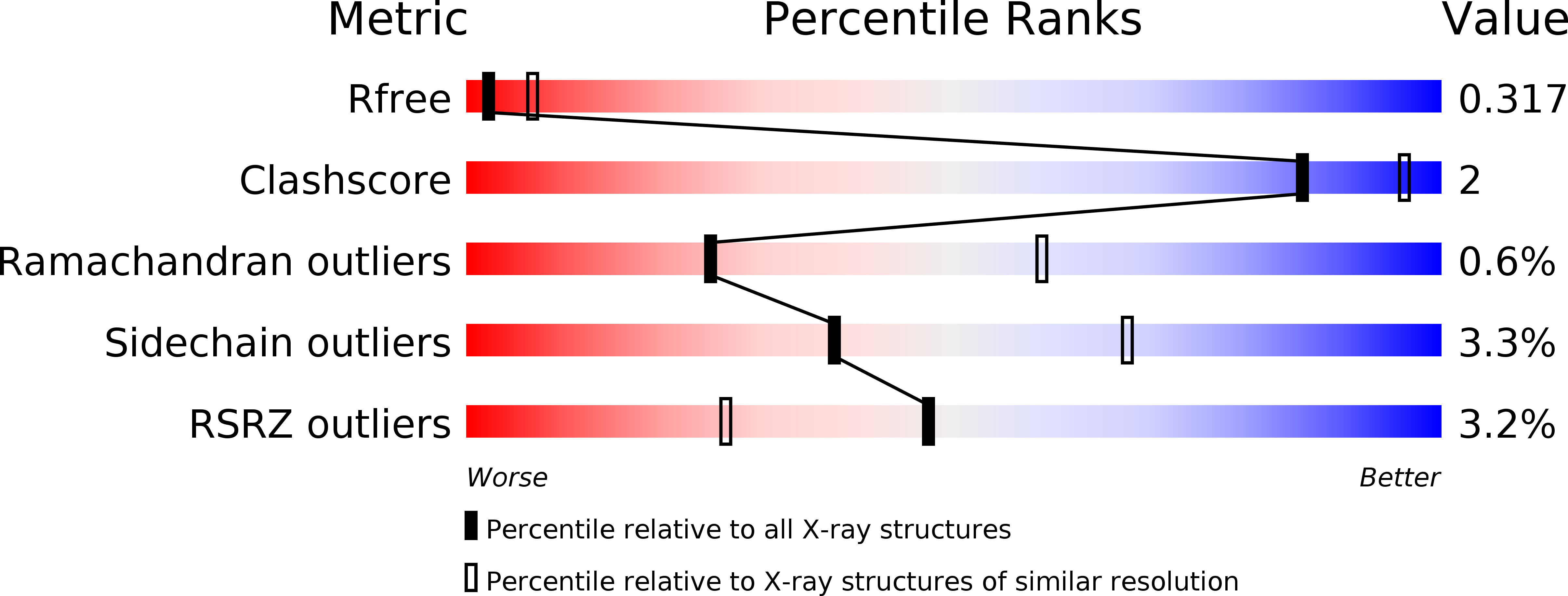
6V8B, 6V8K, 6V8N, 6V90 - PubMed Abstract:
EP300 and CBP (KAT3A/3B) are two highly homologous, multidomain, epigenetic coregulators that play central roles in transcription through the acetylation of lysine residues on histones and other proteins. Both enzymes have been implicated in human diseases, especially cancer. From a high-throughput screen of 191 000 compounds searching for EP300/CBP histone acetyltransferase (HAT) inhibitors, 18 compounds were characterized by a suite of biochemical enzymatic assays and biophysical methods, including X-ray crystallography and native mass spectrometry. This work resulted in the discovery of three distinct mechanistic classes of EP300/CBP HAT inhibitors, including two classes not previously described. The profiles of an example of each class of inhibitor are described in detail. A subsequent medicinal chemistry effort led to the development of a novel class of orally bioavailable AcCoA-competitive EP300/CBP HAT inhibitors with in vivo activity. We believe that this work will prove to be a useful guide for other groups interested in the development of HAT inhibitors.
Organizational Affiliation:
Constellation Pharmaceuticals, 215 First Street, Cambridge, MA 02142.