Elucidation of an Allosteric Mode of Action for a Thienopyrazole ROR gamma t Inverse Agonist.
de Vries, R.M.J.M., Doveston, R.G., Meijer, F.A., Brunsveld, L.(2020) ChemMedChem 15: 561-565
- PubMed: 32053744
- DOI: https://doi.org/10.1002/cmdc.202000044
- Primary Citation of Related Structures:
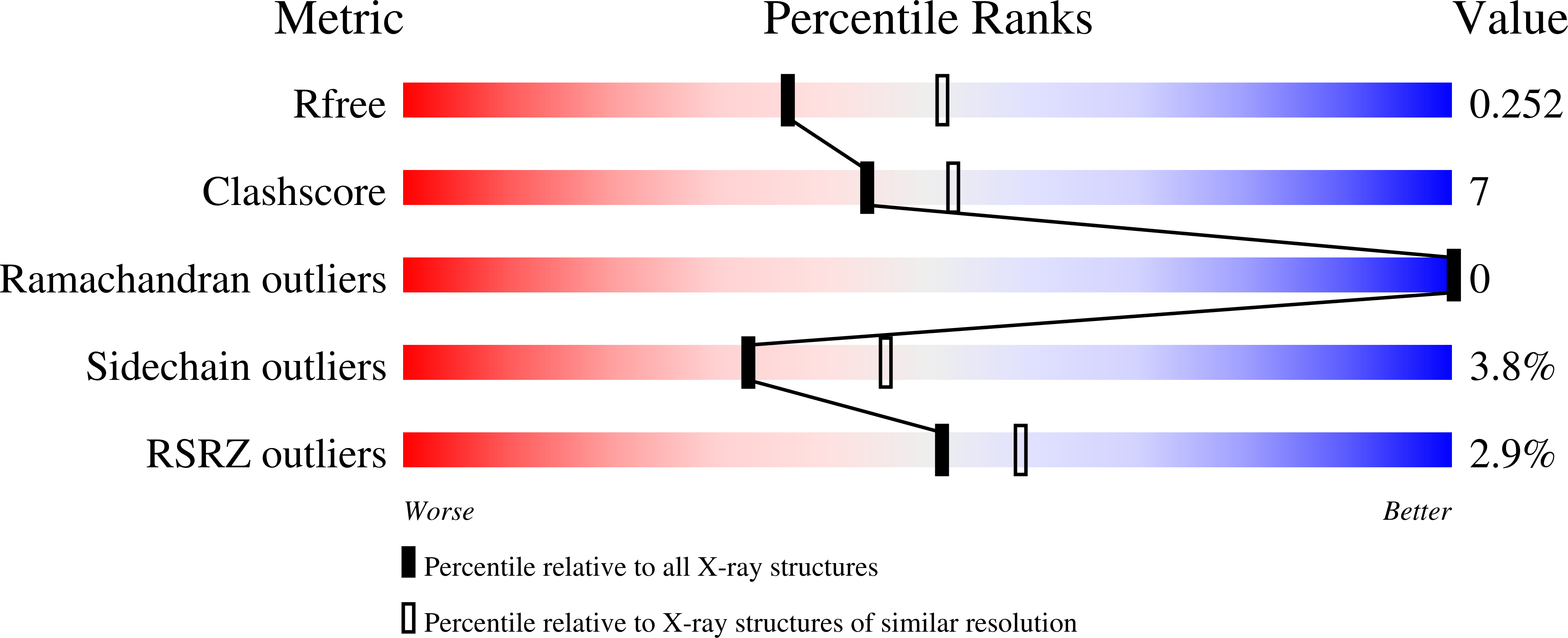
6TLM - PubMed Abstract:
The demand for allosteric targeting of nuclear receptors is high, but examples are limited, and structural information is scarce. The retinoic acid-related orphan receptor gamma t (RORγt) is an important transcriptional regulator for the differentiation of T helper 17 cells for which the first, and some of the most promising, examples of allosteric nuclear receptor modulation have been reported and structurally proven. In a 2015 patent, filed by the pharmaceutical company Glenmark, a new class of small molecules was reported that act as potent inverse agonists for RORγt. A compound library around the central thienopyrazole scaffold captured a clear structure-activity relationship, but the binding mechanism of this new class of RORγt modulators has not been elucidated. Using a combination of biochemical and X-ray crystallography studies, here the allosteric mechanism for the inverse agonism for the most potent compound, classified in the patent as "example 13", is reported, providing a strongly desired additional example of allosteric nuclear receptor targeting.
Organizational Affiliation:
Department of Biomedical Engineering and Institute for Complex Molecular Systems, Eindhoven University of Technology, Den Dolech 2, 5612 AZ, Eindhoven, The Netherlands.