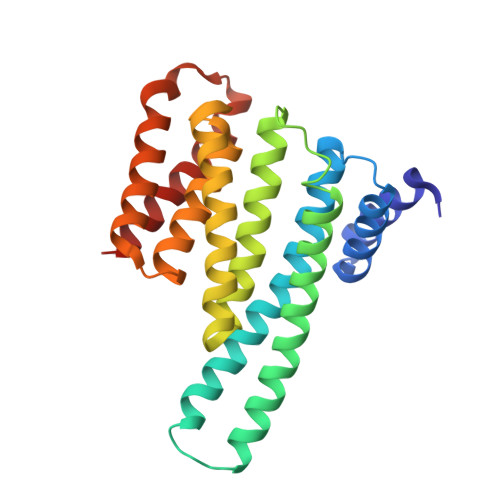
14-3-3 protein binding blocks the dimerization interface of caspase-2.
Kalabova, D., Filandr, F., Alblova, M., Petrvalska, O., Horvath, M., Man, P., Obsil, T., Obsilova, V.(2020) FEBS J 287: 3494-3510
- PubMed: 31961068
- DOI: https://doi.org/10.1111/febs.15215
- Primary Citation of Related Structures:
6S9K, 6SAD - PubMed Abstract:
Among all species, caspase-2 (C2) is the most evolutionarily conserved caspase required for effective initiation of apoptosis following death stimuli. C2 is activated through dimerization and autoproteolytic cleavage and inhibited through phosphorylation at Ser 139 and Ser 164 , within the linker between the caspase recruitment and p19 domains of the zymogen, followed by association with the adaptor protein 14-3-3, which maintains C2 in its immature form procaspase (proC2). However, the mechanism of 14-3-3-dependent inhibition of C2 activation remains unclear. Here, we report the structural characterization of the complex between proC2 and 14-3-3 by hydrogen/deuterium mass spectrometry and protein crystallography to determine the molecular basis for 14-3-3-mediated inhibition of C2 activation. Our data reveal that the 14-3-3 dimer interacts with proC2 not only through ligand-binding grooves but also through other regions outside the central channel, thus explaining the isoform-dependent specificity of 14-3-3 protein binding to proC2 and the substantially higher binding affinity of 14-3-3 protein to proC2 than to the doubly phosphorylated peptide. The formation of the complex between 14-3-3 protein and proC2 does not induce any large conformational change in proC2. Furthermore, 14-3-3 protein interacts with and masks both the nuclear localization sequence and the C-terminal region of the p12 domain of proC2 through transient interactions in which both the p19 and p12 domains of proC2 are not firmly docked onto the surface of 14-3-3. This masked region of p12 domain is involved in C2 dimerization. Therefore, 14-3-3 protein likely inhibits proC2 activation by blocking its dimerization surface. DATABASES: Structural data are available in the Protein Data Bank under the accession numbers 6SAD and 6S9K.
Organizational Affiliation:
Division BIOCEV, Department of Structural Biology of Signaling Proteins, Institute of Physiology of the Czech Academy of Sciences, Vestec, Czech Republic.