Structure of human excitatory amino acid transporter 3 (EAAT3)
Baronina, A., Pike, A.C.W., Han, S., Carpenter, E.P.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Excitatory amino acid transporter 3 | 487 | Homo sapiens | Mutation(s): 2 Gene Names: SLC1A1, EAAC1, EAAT3 Membrane Entity: Yes | 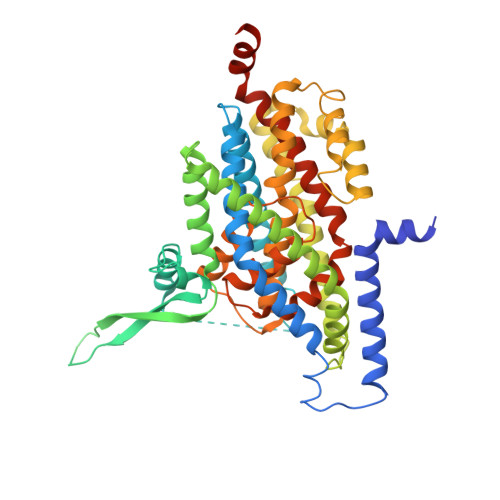 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P43005 (Homo sapiens) Explore P43005 Go to UniProtKB: P43005 | |||||
PHAROS: P43005 GTEx: ENSG00000106688 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P43005 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PC1 Query on PC1 | F [auth A] G [auth A] H [auth A] I [auth A] J [auth A] | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE C44 H88 N O8 P NRJAVPSFFCBXDT-HUESYALOSA-N |  | ||
| Y01 Query on Y01 | E [auth A], M [auth B], T [auth C] | CHOLESTEROL HEMISUCCINATE C31 H50 O4 WLNARFZDISHUGS-MIXBDBMTSA-N |  | ||
| 7O9 (Subject of Investigation/LOI) Query on 7O9 | D [auth A], L [auth B], S [auth C] | (2~{S},3~{S})-2-azanyl-3-[[3-[[4-(trifluoromethyl)phenyl]carbonylamino]phenyl]methoxy]butanedioic acid C19 H17 F3 N2 O6 LPWONNPEPDHEAI-GJZGRUSLSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | RELION | 3.0 |
| MODEL REFINEMENT | PHENIX | 1.14 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Wellcome Trust | United Kingdom | 106169/Z/14/Z |
| Biotechnology and Biological Sciences Research Council | United Kingdom | -- |