Sub-Atomic Resolution Crystal Structures Reveal Conserved Geometric Outliers at Functional Sites.
Laulumaa, S., Kursula, P.(2019) Molecules 24
- PubMed: 31443388
- DOI: https://doi.org/10.3390/molecules24173044
- Primary Citation of Related Structures:
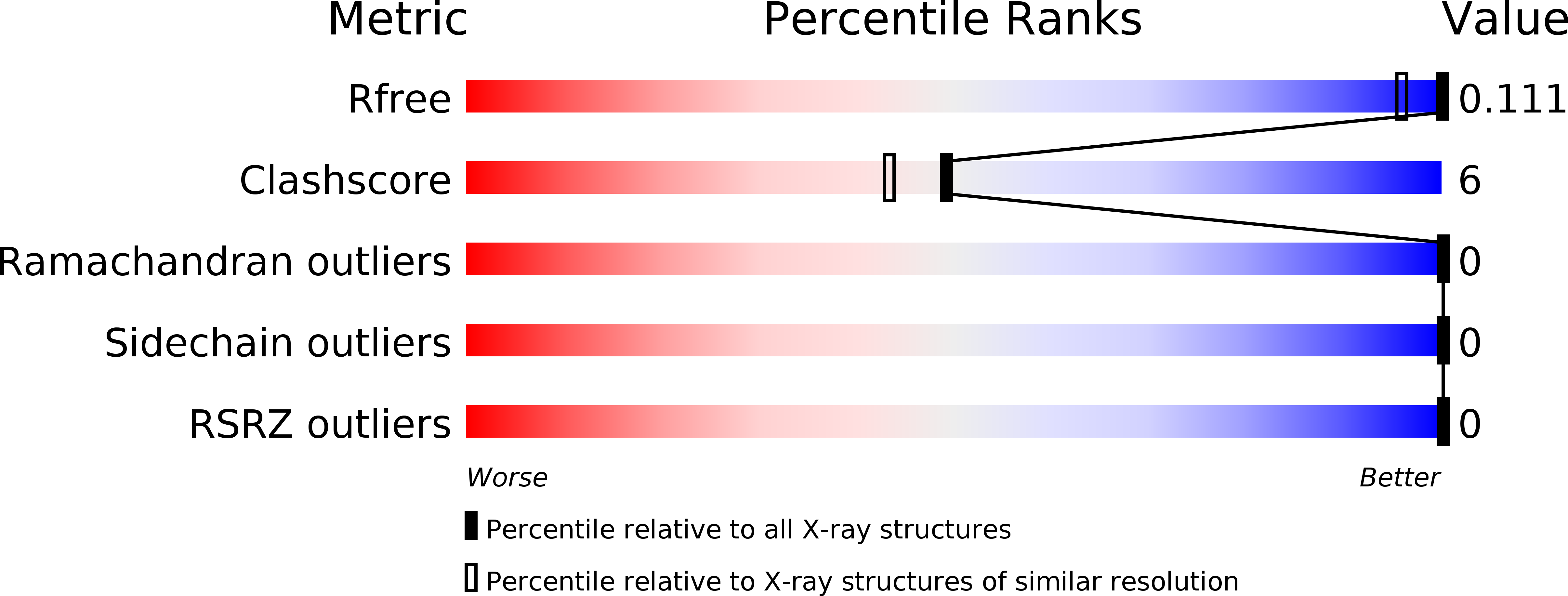
6S2M, 6S2S - PubMed Abstract:
Myelin protein 2 (P2) is a peripheral membrane protein of the vertebrate nervous system myelin sheath, having possible roles in both lipid transport and 3D molecular organization of the multilayered myelin membrane. We extended our earlier crystallographic studies on human P2 and refined its crystal structure at an ultrahigh resolution of 0.72 Å in perdeuterated form and 0.86 Å in hydrogenated form. Characteristic differences in C-H…O hydrogen bond patterns were observed between extended β strands, kinked or ending strands, and helices. Often, side-chain C-H groups engage in hydrogen bonding with backbone carbonyl moieties. The data highlight several amino acid residues with unconventional conformations, including both bent aromatic rings and twisted guanidinium groups on arginine side chains, as well as non-planar peptide bonds. In two locations, such non-ideal conformations cluster, providing proof of local functional strain. Other ultrahigh-resolution protein structures similarly contain chemical groups, which break planarity rules. For example, in Src homology 3 (SH3) domains, a conserved bent aromatic residue is observed near the ligand binding site. Fatty acid binding protein (FABP) 3, belonging to the same family as P2, has several side chains and peptide bonds bent exactly as those in P2. We provide a high-resolution snapshot on non-ideal conformations of amino acid residues under local strain, possibly relevant to biological function. Geometric outliers observed in ultrahigh-resolution protein structures are real and likely relevant for ligand binding and conformational changes. Furthermore, the deuteration of protein and/or solvent are promising variables in protein crystal optimization.
Organizational Affiliation:
Faculty of Biochemistry and Molecular Medicine & Biocenter Oulu, University of Oulu, 90014 Oulu, Finland.