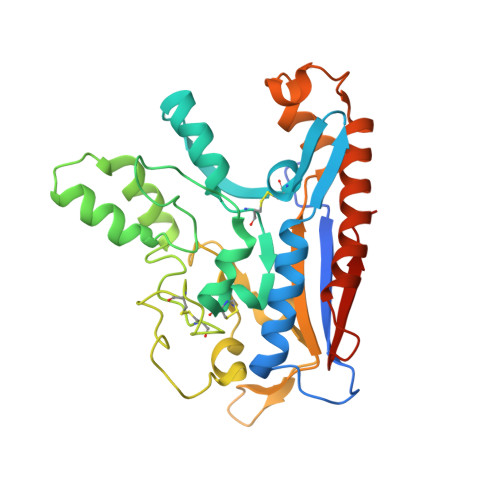
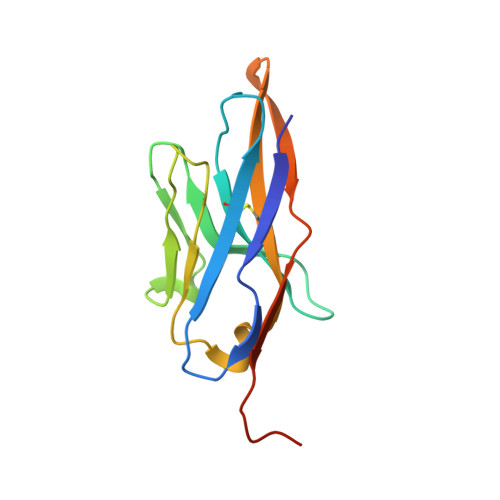
The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Rudolf, A.F., Kinnebrew, M., Kowatsch, C., Ansell, T.B., El Omari, K., Bishop, B., Pardon, E., Schwab, R.A., Malinauskas, T., Qian, M., Duman, R., Covey, D.F., Steyaert, J., Wagner, A., Sansom, M.S.P., Rohatgi, R., Siebold, C.(2019) Nat Chem Biol 15: 975-982
- PubMed: 31548691
- DOI: https://doi.org/10.1038/s41589-019-0370-y
- Primary Citation of Related Structures:
6RTW, 6RTX, 6RTY, 6RVC, 6RVD - PubMed Abstract:
Hedgehog (HH) ligands, classical morphogens that pattern embryonic tissues in all animals, are covalently coupled to two lipids-a palmitoyl group at the N terminus and a cholesteroyl group at the C terminus. While the palmitoyl group binds and inactivates Patched 1 (PTCH1), the main receptor for HH ligands, the function of the cholesterol modification has remained mysterious. Using structural and biochemical studies, along with reassessment of previous cryo-electron microscopy structures, we find that the C-terminal cholesterol attached to Sonic hedgehog (Shh) binds the first extracellular domain of PTCH1 and promotes its inactivation, thus triggering HH signaling. Molecular dynamics simulations show that this interaction leads to the closure of a tunnel through PTCH1 that serves as the putative conduit for sterol transport. Thus, Shh inactivates PTCH1 by grasping its extracellular domain with two lipidic pincers, the N-terminal palmitate and the C-terminal cholesterol, which are both inserted into the PTCH1 protein core.
Organizational Affiliation:
Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, Oxford, UK.