Staphylococcal Complement Evasion Protein Sbi Stabilises C3d Dimers by Inducing an N-Terminal Helix Swap
Dunphy, R.W., Wahid, A.A., Back, C.R., Martin, R.L., Watts, A.G., Dodson, C.A., Crennell, S.J., van den Elsen, J.M.H.(2022) Front Immunol 13
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2022) Front Immunol 13
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Complement C3 | A, C [auth B] | 310 | Homo sapiens | Mutation(s): 0 Gene Names: C3, CPAMD1 | 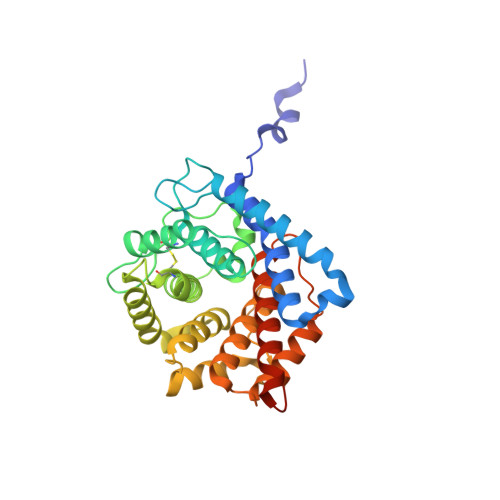 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P01024 (Homo sapiens) Explore P01024 Go to UniProtKB: P01024 | |||||
PHAROS: P01024 GTEx: ENSG00000125730 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P01024 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Immunoglobulin-binding protein Sbi | B [auth C], D | 81 | Staphylococcus aureus subsp. aureus Mu50 | Mutation(s): 0 Gene Names: sbi, SAV2418 |  |
UniProt | |||||
Find proteins for Q2FVK5 (Staphylococcus aureus (strain NCTC 8325 / PS 47)) Explore Q2FVK5 Go to UniProtKB: Q2FVK5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q2FVK5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PEG Query on PEG | E [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | F [auth B] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 73.518 | α = 90 |
| b = 115.169 | β = 90 |
| c = 118.925 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| DIALS | data reduction |
| Aimless | data scaling |
| BALBES | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Biotechnology and Biological Sciences Research Council | United Kingdom | BB/N022165/1 |