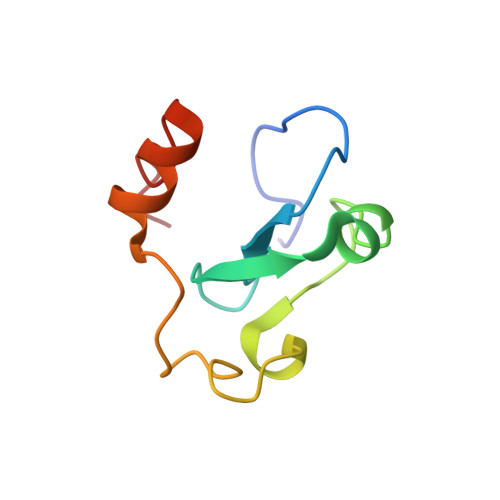
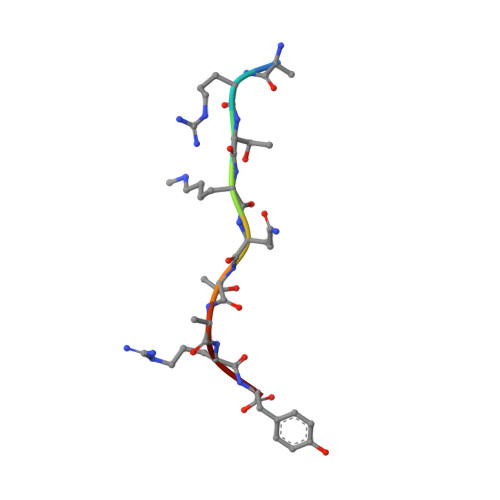
The Arabidopsis (ASHH2) CW domain binds monomethylated K4 of the histone H3 tail through conformational selection.
Dobrovolska, O., Brilkov, M., Madeleine, N., Odegard-Fougner, O., Stromland, O., Martin, S.R., De Marco, V., Christodoulou, E., Teigen, K., Isaksson, J., Underhaug, J., Reuter, N., Aalen, R.B., Aasland, R., Halskau, O.(2020) FEBS J 287: 4458-4480
- PubMed: 32083791
- DOI: https://doi.org/10.1111/febs.15256
- Primary Citation of Related Structures:
6QXZ - PubMed Abstract:
Chromatin post-translational modifications are thought to be important for epigenetic effects on gene expression. Methylation of histone N-terminal tail lysine residues constitutes one of many such modifications, executed by families of histone lysine methyltransferase (HKMTase). One such protein is ASHH2 from the flowering plant Arabidopsis thaliana, equipped with the interaction domain, CW, and the HKMTase domain, SET. The CW domain of ASHH2 is a selective binder of monomethylation at lysine 4 on histone H3 (H3K4me1) and likely helps the enzyme dock correctly onto chromatin sites. The study of CW and related interaction domains has so far been emphasizing lock-key models, missing important aspects of histone-tail CW interactions. We here present an analysis of the ASHH2 CW-H3K4me1 complex using NMR and molecular dynamics, as well as mutation and affinity studies of flexible coils. β-augmentation and rearrangement of coils coincide with changes in the flexibility of the complex, in particular the η1, η3 and C-terminal coils, but also in the β1 and β2 strands and the C-terminal part of the ligand. Furthermore, we show that mutating residues with outlier dynamic behaviour affect the complex binding affinity despite these not being in direct contact with the ligand. Overall, the binding process is consistent with conformational selection. We propose that this binding mechanism presents an advantage when searching for the correct post-translational modification state among the highly modified and flexible histone tails, and also that the binding shifts the catalytic SET domain towards the nucleosome. DATABASES: Structural data are available in the PDB database under the accession code 6QXZ. Resonance assignments for CW42 in its apo- and holo-forms are available in the BMRB database under the accession code 27251.
Organizational Affiliation:
Department of Biological Sciences, University of Bergen, Norway, Bergen.