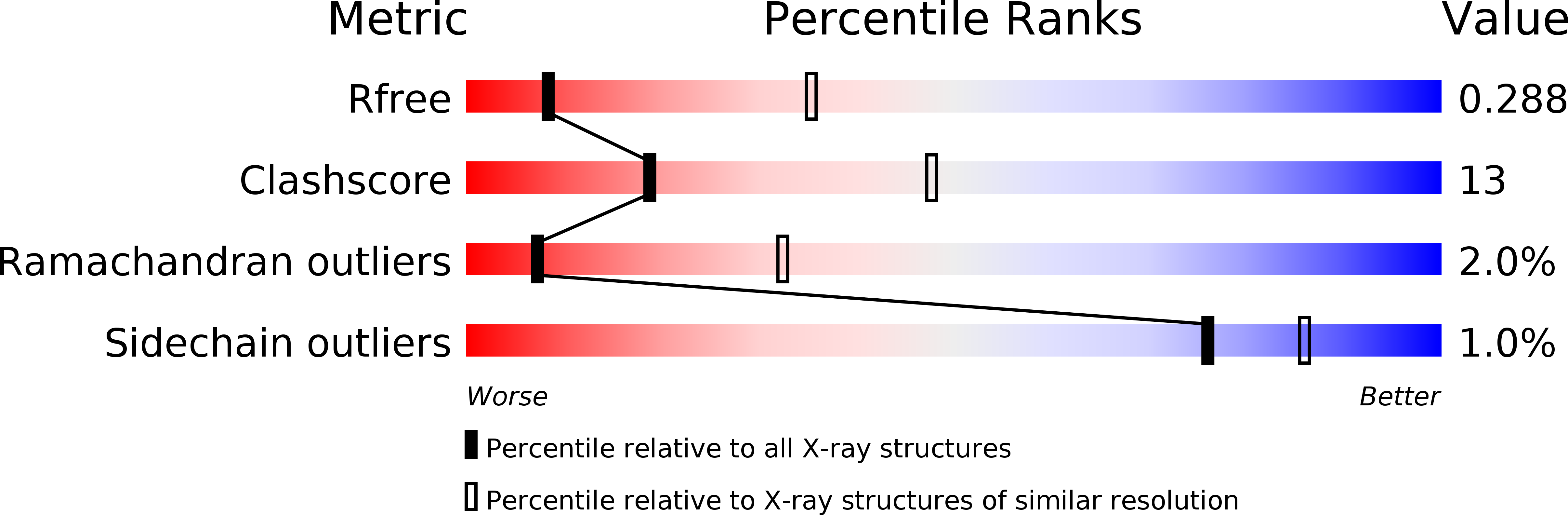
In vivo efficacy of mutant IDH1 inhibitor HMS-101 and structural resolution of distinct binding site.
Chaturvedi, A., Goparaju, R., Gupta, C., Weder, J., Klunemann, T., Araujo Cruz, M.M., Kloos, A., Goerlich, K., Schottmann, R., Othman, B., Struys, E.A., Bahre, H., Grote-Koska, D., Brand, K., Ganser, A., Preller, M., Heuser, M.(2020) Leukemia 34: 416-426
- PubMed: 31586149
- DOI: https://doi.org/10.1038/s41375-019-0582-x
- Primary Citation of Related Structures:
6Q6F - PubMed Abstract:
Mutations in isocitrate dehydrogenase 1 (IDH1) are found in 6% of AML patients. Mutant IDH produces R-2-hydroxyglutarate (R-2HG), which induces histone- and DNA-hypermethylation through the inhibition of epigenetic regulators, thus linking metabolism to tumorigenesis. Here we report the biochemical characterization, in vivo antileukemic effects, structural binding, and molecular mechanism of the inhibitor HMS-101, which inhibits the enzymatic activity of mutant IDH1 (IDH1mut). Treatment of IDH1mut primary AML cells reduced 2-hydroxyglutarate levels (2HG) and induced myeloid differentiation in vitro. Co-crystallization of HMS-101 and mutant IDH1 revealed that HMS-101 binds to the active site of IDH1mut in close proximity to the regulatory segment of the enzyme in contrast to other IDH1 inhibitors. HMS-101 also suppressed 2HG production, induced cellular differentiation and prolonged survival in a syngeneic mutant IDH1 mouse model and a patient-derived human AML xenograft model in vivo. Cells treated with HMS-101 showed a marked upregulation of the differentiation-associated transcription factors CEBPA and PU.1, and a decrease in cell cycle regulator cyclin A2. In addition, the compound attenuated histone hypermethylation. Together, HMS-101 is a unique inhibitor that binds to the active site of IDH1mut directly and is active in IDH1mut preclinical models.
Organizational Affiliation:
Department of Hematology, Hemostasis, Oncology and Stem Cell Transplantation, Hannover Medical School, Hannover, Germany.